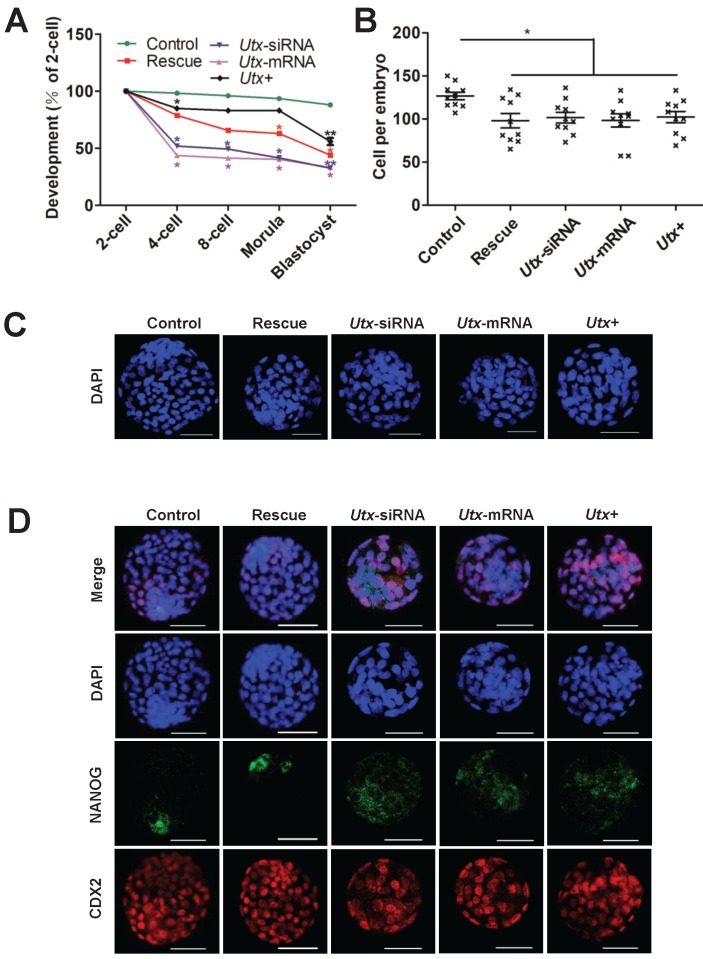
Figure 4.
Fine-tuning expression of Zscan4d partially rescues si-Utx embryonic development. (A) Developmental rates in the rescued (Utx-siRNA+Zscan4d-mRNA) and other groups (Control, Utx-siRNA, Utx-mRNA, Utx+) of mouse embryos cultured in vitro. The efficiency was calculated based on the number of 2-cell embryos. Error bars indicate SD, n ≥ 3. *P < 0.05, ** P < 0.01 by the two-tailed Student's t-test. (B) and (C) Cell numbers of blastocyst determined by the nuclei stained by DAPI and representative DAPI staining of blastocysts of the rescue (Utx-siRNA+Zscan4d-mRNA) and other groups (Control, Utx-siRNA, Utx-mRNA, Utx+) of mouse embryos after 120 h of culture in vitro. Error bars indicate SEM. *P < 0.05 by the two-tailed Student's t-test. Scale bar, 50 μm. (D) Immunofluorescence images of the rescue (Utx-siRNA+Zscan4d-mRNA) and other groups (Utx-siRNA, Utx-mRNA, Utx+) of mouse embryos after 120 h of culture in vitro. NANOG (ICM) and CDX2 (TE) were used as lineage markers. Representative images from ≥ 20 embryos analyzed in independent micromanipulations for each condition are shown. Scale bar, 50 μm.