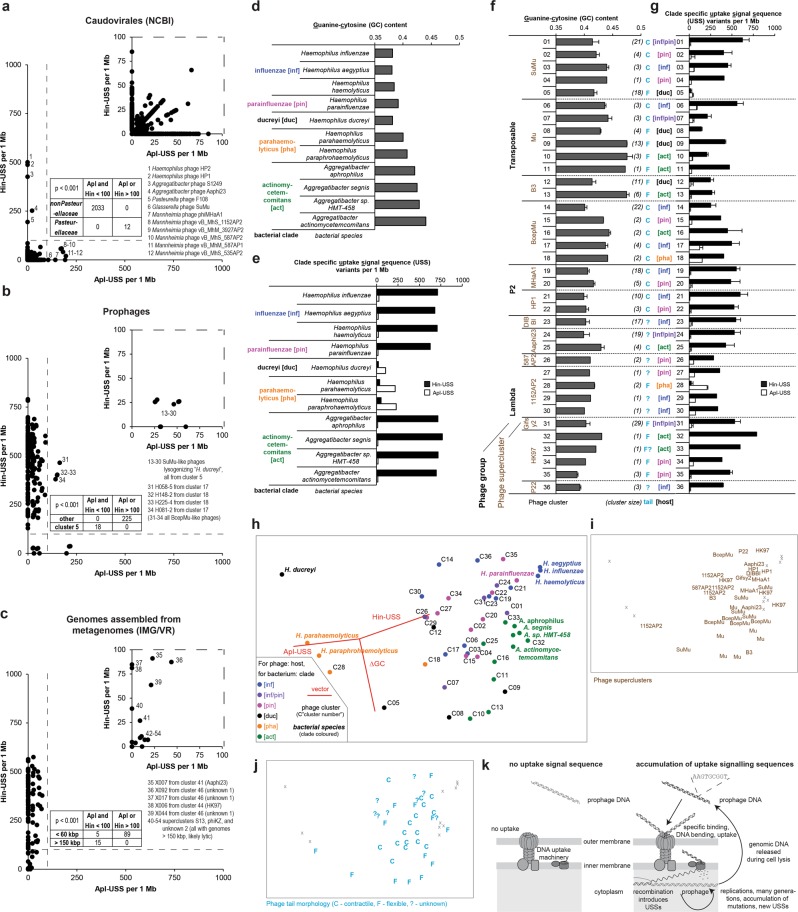
Fig. 7.
Frequencies of DNA uptake signal sequences (USSs) and guanine–cytosine content (GC%) in genome sequences of Pasteurellaceae species and their phages. Frequencies of clade-specific uptake signal sequence (USS) variants in both orientations and guanine–cytosine content (GC%) were measured for genomes from diverse phages and human-associated Pasteurellaceae species. Frequencies are given per 1 Mb (i.e., 106 nt) of genome sequence. a Scatter plot showing frequencies of H. influenzae USS (Hin-USS) and Actinobacillus pleuropneumoniae USS (Apl-USS) in genomes of phages from order Caudovirales [25] on y- and x-axis, respectively. Enlarged fragment of plot covering low values is shown in top right corner. Contingency table summarizing studied groups is shown in the middle. Fisher’s exact test (two-sided) was used to analyze the significance of the association between the phage groups and presence of USSs at given cutoff. Phages characterized by noteworthy values are numbered and labeled. b like a but prophages classified in this study are presented. c like a but IMG/VR assemblies [27] are presented. d GC content was depicted for selected species grouped in clades based on genetic relationship inferred from concatenated nucleotide sequences (~2650 nt) of 16 S rRNA and three housekeeping (infB, pgi, recA) genes [1]. The subfamily clade labels were colored and abbreviated in square brackets throughout the figure. Genome sequences from either the representative National Center for Biotechnology Information (NCBI) strains or the type strains were retrieved from NCBI genome database. e Frequencies of Hin- and Apl-USS in the same genomes as in d. f GC content (mean ± s.d.) was depicted for phage clusters gathered in groups and superclusters (latter in brown throughout the figure). g Frequencies (mean ± s.d., per 1 Mb) of Hin-USS and Apl-USS in prophage sequences grouped like in f. For clusters depicted in f and g additional information is provided: the cluster size, which is the number of studied phages and written in parentheses, the predicted or confirmed morphology of phage tail written in light blue throughout the figure (C—contractile, F—flexible,?—unknown), and the abbreviated clade name for the phage host. h Ordination of bacterial species and phage clusters was constructed based on GC% and USS frequencies from d to g. The Bray-Curtis coefficient was calculated between every pair of samples using three variables: ΔGC (i.e., GC content reduced by the minimal GC in studied dataset), frequencies of Hin-USS, frequencies of Apl-USSs, each standardized by maximum (i.e., values were scaled so that their maxima across these three variables were always 100). Non-metric multidimensional scaling (nMDS) was used to represent the samples in two-dimensional space. Points were colored based on bacterial clades and phage host (labels starting with “C”). Superimposed is a vector plot for three variables (in red), with the vector direction for each variable reflecting the Pearson correlations of their values with the ordination axes, and length giving the multiple correlation coefficient from this linear regression on the ordination points. 2D-stress of 0.12 was observed. Same ordination was used in i and j. i Supercluster assignment was plotted for all phage clusters. Location of bacterial species is indicated by gray “x”. j Phage tail morphology for all phage clusters is given. k Mechanism of USS accumulation in prophages. Picture adapted from [52]