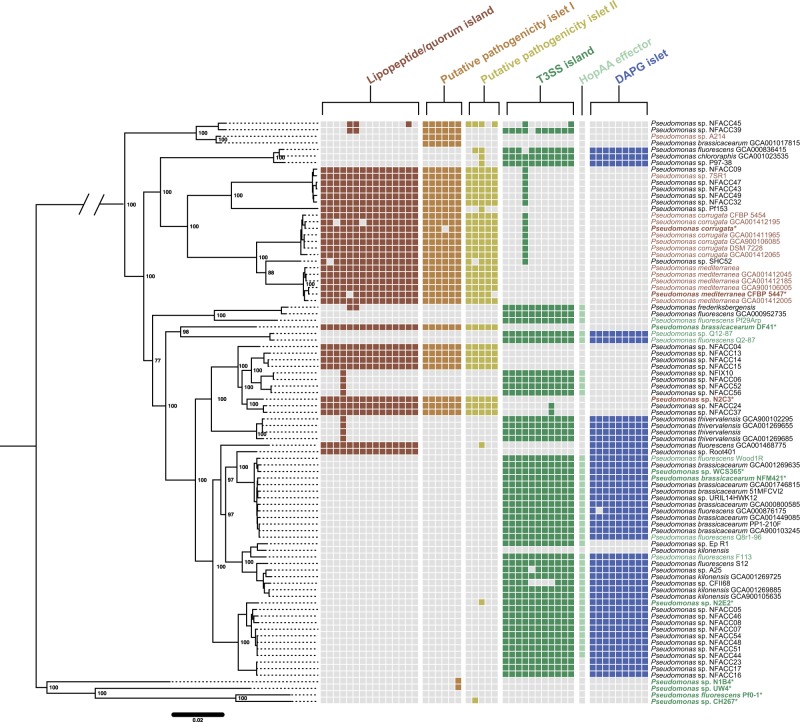
Fig. 3.
Polyphyletic distribution of pathogenic and commensal genomic islands within the bcm clade. A species tree was built for the bcm clade using a concatenated alignment of 2030 conserved genes. Color-coded squares represent presence and absence of individual genes associated with each locus based on PyParanoid presence-absence data. In the case of both the LPQ and T3SS, only a subset of genes to the island are shown because both islands contain genes that are part of larger homolog groups not specific to either island. Bold taxon names highlight strains experimentally tested for commensal (green) or pathogenic (red) behavior (Fig. 2g). For strains that we did not test experimentally, we annotated rhizosphere lifestyle based on previous reports in the literature with the same color scheme. All six loci exhibit a highly polyphyletic distribution in extant strains suggesting that the pathogenic and commensal lifestyles have a complex evolutionary history in this clade