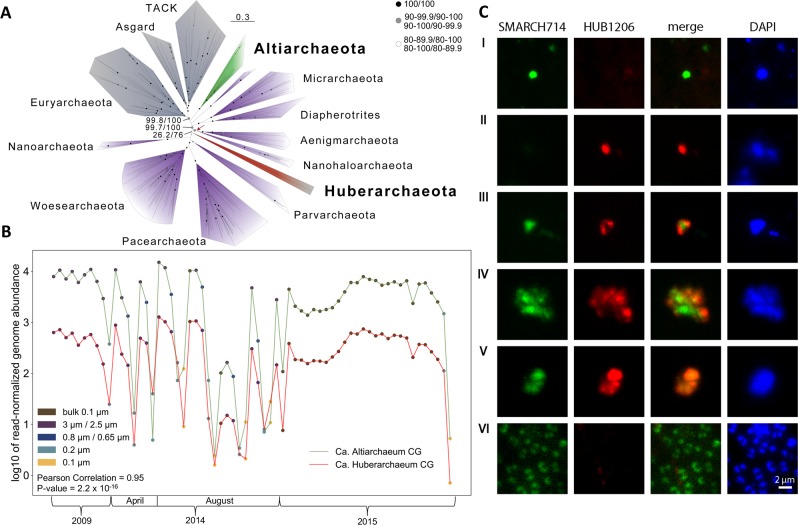
Fig. 1.
Phylogeny and co-occurrence analysis of Ca. Huberiarchaeum crystalense and Ca. Altiarchaeum hamiconexum CG. a Phylogenetic placement of Huberarchaeota using a concatenated protein alignment of 34 marker genes (DPANN archaea are in purple). This maximum likelihood tree includes 186 taxa and was inferred based on an alignment of 4224 positions in IQ-tree under the LG + C60 + F + R model. The red arrow highlights the alternative placement of the Huberarchaeota in the IQ-tree analyses based on an SR4 recoded alignment (support: 6.2/76). Bootstrap support was inferred using an SH-like approximate likelihood ratio test and ultrafast bootstrap method. Black circle: 100% support for both SH and ultrafast bootstrap methods. Gray circle: Support values from 90–99.9 for eithermethod and including branches were only one bootstrap method had a support value of 100. White circle: Support values from 80–89.9 for either method and including branches were only one bootstrap method had a support value of 90. Scale bar indicates the average number of substitutions per site. For details on the methods please see supplementary data. b Co-correlation analysis of relative abundance of genomes of Ca. H. crystalense and Ca. A. hamiconexum CG based on 65 metagenome samples taken between 2009 and 2015. The co-occurrence also holds true between samples taken with filters of different pore-sizes indicating that the organisms form an association that does not pass through small pore sizes. A detailed analysis of the proportionality of 505 different organisms from the ecosystems is provided in Fig. S2. c MiL fluorescence in situ hybridization depicting the association of Ca. H. crystalense and Ca. A. hamiconexum CG cells and the respective controls. SMARCH714 is a specific probe for Altiarchaeota [11] labeled with Atto488. HUB1206 is a probe designed off the 16S rRNA gene sequence of Ca. H. crystalense based on information from multiple genomic bins (see Supplementary Methods). The probe was labeled with CY3. I. Individual Ca. A. hamiconexum CG cell. II. Individual Ca. H. crystalense cell. III. Attachment of Ca. H. crystalense to Ca. A. hamiconexum CG IV. occurrence of Ca. H. crystalense to Ca. A. hamiconexum CG in cell clusters V. Overlapping fluorescence signals of the two probes putatively in the same cells. VI. Control sample of a well-studied ecosystem dominated by Ca. A. hamiconexum IMS [19], where according to metagenomic data no Huberarchaeota occur. We show that the specific probe of Ca. H. crystalense does not bind to Altiarchaeota or other organisms in the ecosystem. For counts on the frequency of the observed associations please see Fig. S4