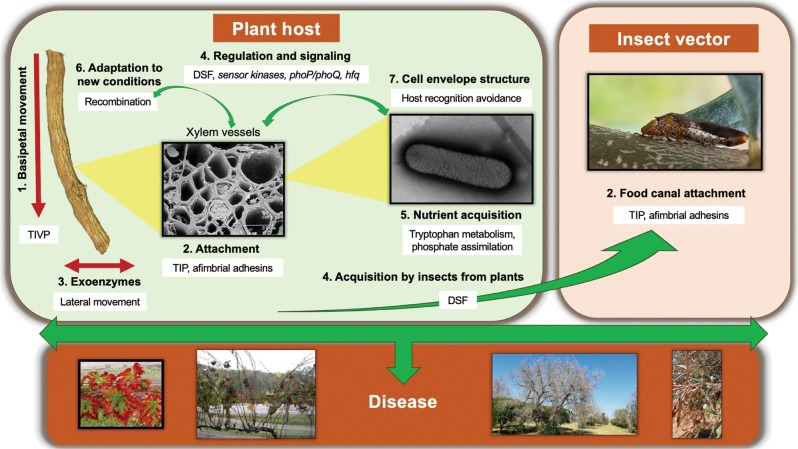
Fig. 7.
Diagram illustrating the ecological role of genes found to be under recent recombination in Xylella fastidiosa. X. fastidiosa is only found in two very specific ecological niches: xylem vessels of host plants and food canal of xylem-feeding insects. Genes that were identified through our analysis as having the highest rate of recent intersubspecific recombination are listed in Table 1, S6, and S7. Based on the functional classification of these genes (Table 1), we placed these functions in the context of the ecology of X. fastidiosa during its life cycle. Briefly, in plant hosts (left panel), X. fastidiosa needs twitching motility [1] and exoenzymes [3] to colonize the xylem system inside plants, where nutrients are very limited [5]. While colonizing xylem vessels, they need attachment [2] to withstand constant liquid flow in the vascular system and to form biofilms. Proteins in the cell envelope are fundamental for recognition by the plant [7]. When colonizing a new host, X. fastidiosa most likely needs to acquire new genetic information for adaptation to new conditions [6]. Several regulatory cascades detected to be under recombination, have been implicated in regulating bacterial colonization and virulence [4]. DSF quorum-sensing molecule has a role in causing an increase in X. fastidiosa attachment and biofilm formation that is important to increase cell stickiness and therefore insect acquisition [4]. In the food canal of the xylem-feeding insect (right panel), attachment by TIP and afimbrial adhesins [2] is important to sustain population under high liquid flow. TIVP: type IV pili; TIP: type I pili; T2SS: type two secretion system; DSF: diffusible signal factor. Numbers of the highlighted X. fastidiosa traits from 1 to 7 correlate to the categories described in Table 1. For more information refer to the text and Table 1. Photo credits: plant host 1, 2: Harvey Hoch (Cornell University); insect vector 2: Phil Brannen (University of Georgia). All other pictures are credited to the authors