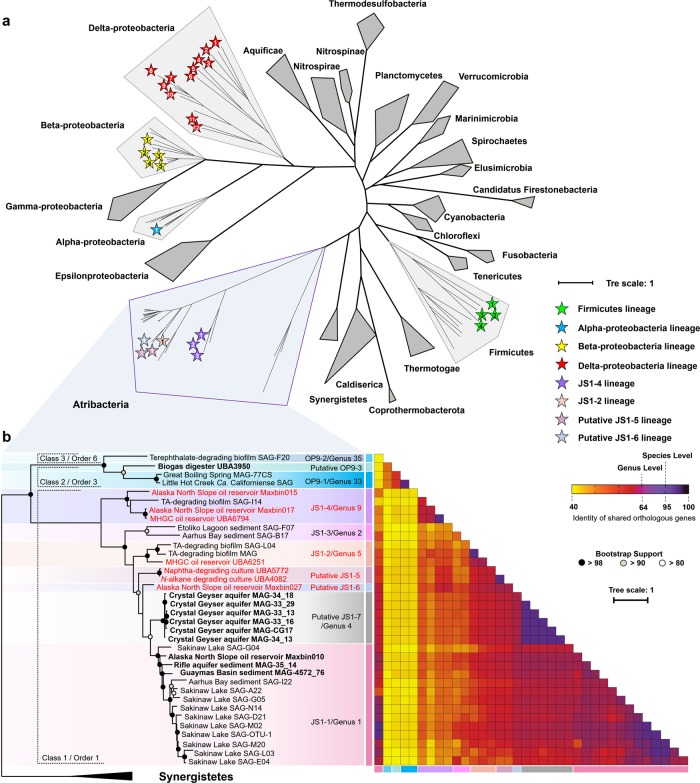
Fig. 1.
Phylogenomic analysis of atribacterial MAGs and SAGs with contamination less than 8%. a Phylogenomic placement of atribacterial genomes in a genome tree constructed with >200 reference archaeal and bacterial genomes. The tree was inferred from the concatenation of 400 conserved marker genes (Supplementary Table S4) [39]. Genomes containing at least one of the fumarate addition genes (ass/bss/nms/hbS) were indicated by stars, and the numbers inside the stars correspond to the taxonomic affiliation in Fig. 2a. b Detail phylogenetic relationships and heatmap of pair-wise orthologous similarity values of Atribacteria OP9 and JS1 as proposed by Nobu et al. [2]. Newly assembled and co-assembled MAGs were indicated in bold, MAGs containing FAE gene were indicated in red. Genes are designated as shared if they are bidirectional BLASTP hits (E-value <10−5). Branches that meet the Genus, Order and Class candidate taxonomic ranks proposed by Yarza et al. [1] are encompassed by dotted parentheses