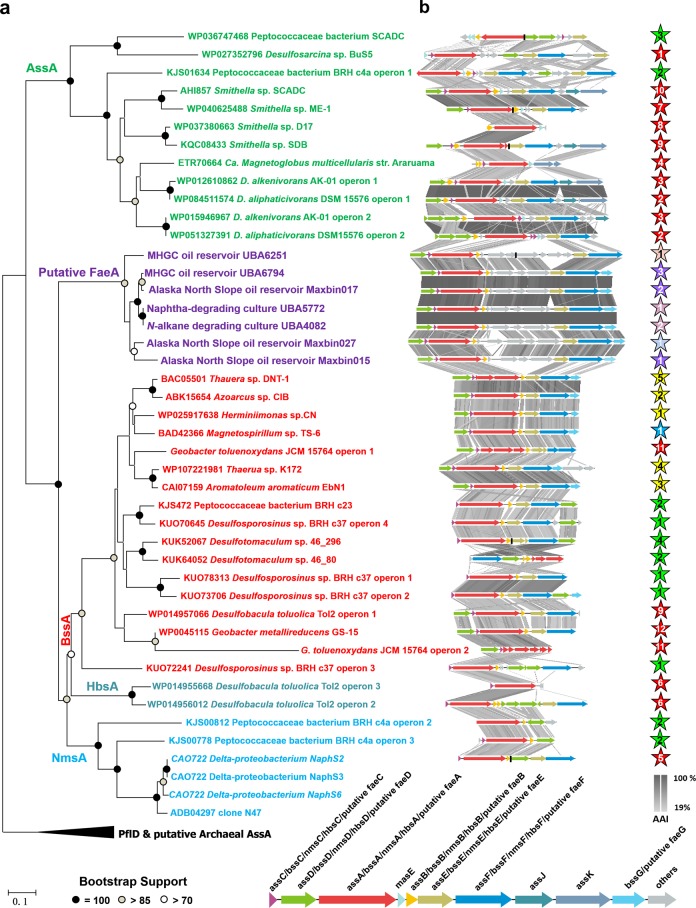
Fig. 2.
Analysis of FAE operons in atribacterial MAGs. a Phylogenetic analysis of Atribacterial faeA (861 amino-acid positions). b TBLASTx comparison of the FAE gene clusters of atribacterial MAGs with all other genomes containing fumarate addition enzymes. For the comparison, an E-value cutoff of 1e−10 was used, and visualization of the gene clusters was done with the program Easyfig [83]. In some genomes, the fumarate addition genes are located in separate contigs due to fragmentary assembly, and in these cases, borders between the single clusters are marked with black vertical lines. The stars on the right side correspond to the branch position of MAGs in Fig. 1a. A representative genomic organization of fumarate addition enzyme genes was shown at the bottom of figure