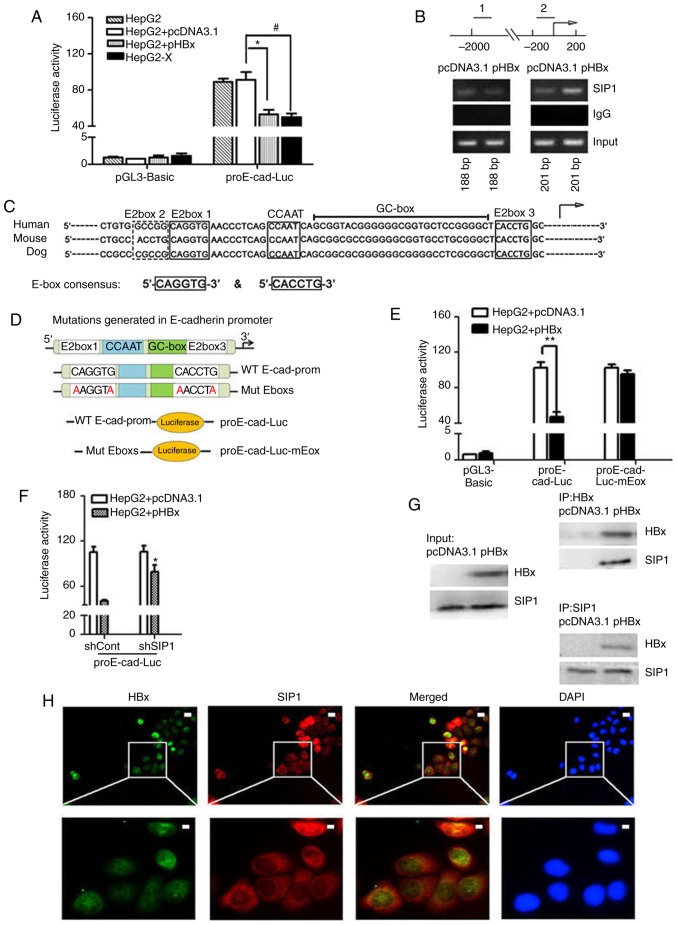
Figure 3.
HBx recruits endogenous SIP1 to the E-cadherin promoter. (A) Dual-luciferase assay of human E-cadherin promoters in Mock treated, pcDNA3.1 or pHBX transfected HepG2 cells and HepG2-X cells. Data were normalized to the luciferase activity of cells treated with pcDNA3.1 and pGL3-Basic. Data are representative of at least three independent experiments. *P<0.05 HepG2 + pcDNA3.1 vs. HepG2 + pHBx; #P<0.05 HepG2 + pcDNA3.1 vs. HepG2-X. (B) ChIP primers were designed on E-cadherin gene regulatory regions. Distal primers correspond to downstream regulatory regions (1) of -1895 to -1707 nt of the E-cadherin gene. The ChIP primers (2) were designed adjacent to the TSS locations across the E-box region of the human E-cadherin promoter. pHBx-transfected HepG2 cell lysates were subjected to ChIP using an anti-SIP1 antibody. PCR was conducted using the indicated primer pairs. An empty vector and IgG served as external and internal negative controls. (C) Sequence homology of the consensus E-box in the E-cadherin promoter of mammalian. E-boxes 1, 3 and 4, CCAAT box and GC box are conserved regulatory elements, as shown in the diagram. The arrow indicates the putative TSS. (D) Mutations generated in the E-boxes carried the E-box 1 mutation CAGGTG → AAGGTA and E-box 3 mutation CACCTG → AACCTA. The wild-type E-cadherin promoter and promoter comprising two mutated E-boxes were cloned into a luciferase vector to construct the proE-cad-Luc and proE-cad-Luc-mEbox plasmids. (E) Dual-luciferase assay of E-cadherin promoter constructs with proE-cad-Luc or proE-cad-Luc-mEbox in pHBx- or pcDNA3.1-transfected HepG2 cells. **P<0.01. (F) Relative E-cadherin promoter activities inshSIP1/shCont and pcDNA3.1/pHBX treated HepG2 cells. *P<0.05; (shSIP1 + WT, vs. shCont + WT). (G) Co-immunoprecipitation in protein extracts of pcDNA3.1-transfected HepG2 cells and HBx-expressing HepG2 cells with anti-SIP1 or anti-HBx antibodies and western blot detection of HBx and SIP1, respectively. (H) Immunofluorescent staining of HepG2-X cells with anti-HBx and anti-SIP1 to show the subcel-lular co-localization of HBx and SIP. DAPI was used to visualize nuclei. Scale bar=10 µm. SIP1, Smad interacting protein 1; HBx, hepatitis B virus X; pHBx, pcDNA3.1-HBx; sh, short hairpin RNA. TSS, transcription start site; WT, proE-cad-Luc; Mut, proE-cad-Luc-mEbox; ChIp, chromatin immunoprecipitation.