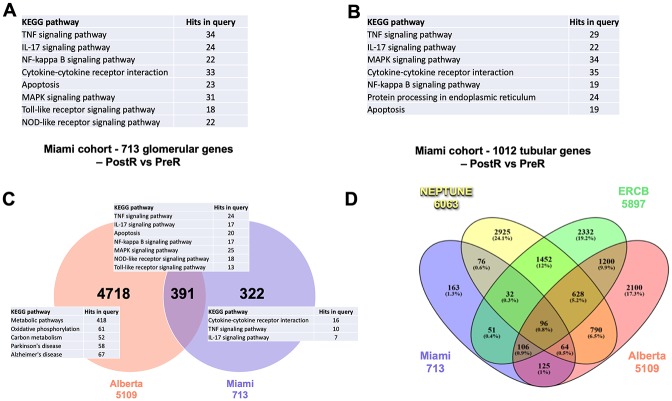
Fig 2. Identification of molecular pathways specifically activated in glomeruli of patients with NON REC FSGS.
Microarray analysis, followed by gene enrichment KEGG pathway analysis of post (PostR) and pre- (PreR) reperfusion kidney biopsy samples obtained from patients with FSGS. (A) 713 genes are differentially regulated in the glomerular compartment of kidney biopsies obtained from patients with FSGS. KEGG pathway analysis identifies dysregulation of predominantly inflammatory as well as apoptotic pathways among the top 20 pathways. Selected pathways out of the top 20 are shown. (B) 1012 genes are differentially regulated in the tubulointerstitial compartment of kidney biopsies obtained from patients with FSGS. KEGG pathway analysis identifies dysregulation of predominantly inflammatory as well as apoptotic pathways among the top 20 pathways. Selected pathways out of the top 20 are shown. (C, D) Identification of differentially expressed genes and pathways specific to FSGS. (C) Glomerular gene expression changes identified in PostR vs PreR biopsies compared to genes differentially expressed in PostR biopsies from 70 kidneys from 53 deceased donors published in the “Alberta” study (GEO accession: GSE37838) are shown. Venn diagram analysis indicating that out of the 713 glomerular genes regulated in PostR vs PreR glomerular biopsies of FSGS patients, 322 genes are uniquely regulated in PostR biopsies of FSGS patients whereas 391 genes are also regulated in kidney biopsies of the Alberta patients. Inflammatory pathways are primarily activated in glomerular biopsies of the Miami cohort, metabolic pathways are particularly activated in the Alberta cohort of patients evaluated for acute kidney injury (C). Venn Diagram analysis demonstrating differentially expressed genes specific to FSGS (D).