Figure 1. RAS-induced H/ACA snoRNAs are required for oncogene-induced senescence in vivo.
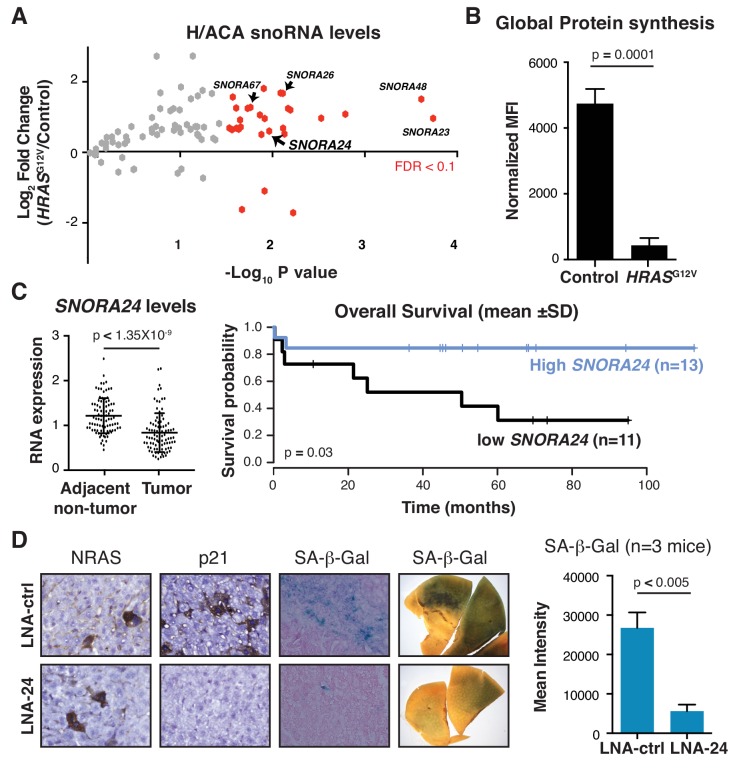
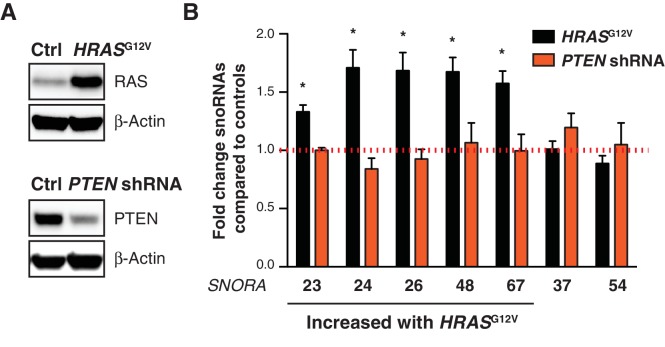
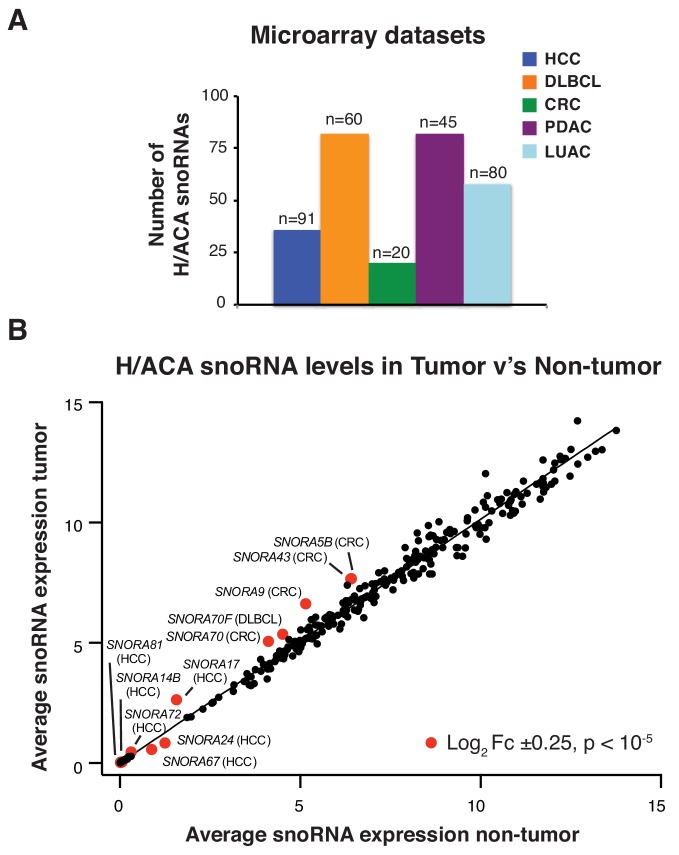
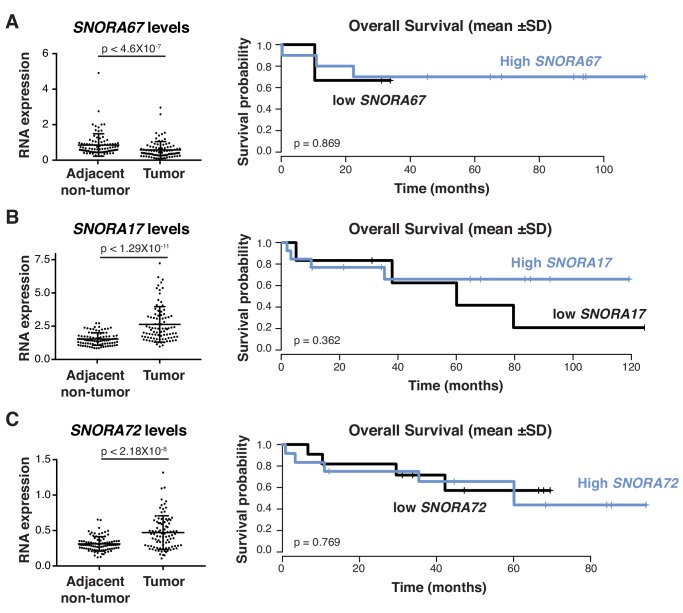
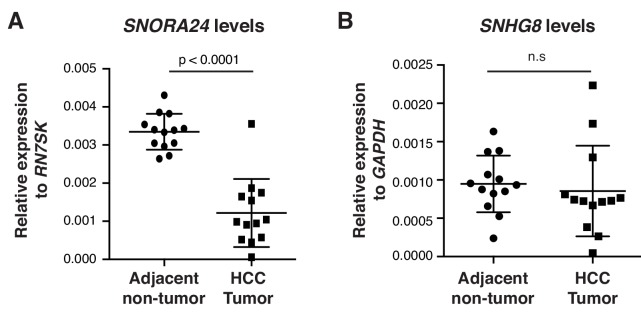
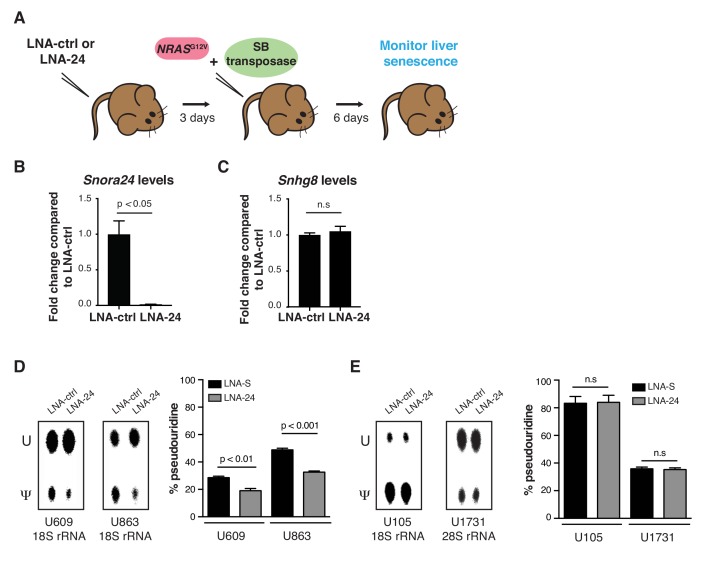
(A) Volcano plot displays Log2 fold change in H/ACA snoRNA levels 5 days following HRASG12V expression in primary human skin fibroblasts measured by snoRNA qPCR array from three independent experiments. SnoRNAs in red exhibit statistically significant fold change in expression in HRASG12V expressing cells compared to controls (p<0.05, unpaired Student’s t-test or FDR < 0.1). SnoRNAs highlighted with labels were independently validated as shown in Figure 1—figure supplement 1B. (B) Graph illustrates mean ± SD mean fluorescent intensity (MFI) of the amount of de novo protein synthesis in primary human fibroblasts 5 days following expression of HRASG12V compared to control treated cells by measuring OPP incorporation into newly synthesized protein from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p=0.0001. (C) Analysis of SNORA24 levels in HCC specimens compared to adjacent non-tumor tissue of 91 HCC patients (GSE25097) (paired Student’s t-test, p<1.35×10−9) (left panel) and Kaplan-Meier curve showing overall survival of HCC patients with high or low SNORA24 levels (mean ± 1 SD of SNORA24 levels) (right panel). Statistical significance was calculated using the log-rank test, with p=0.03. (D) Representative image for NRAS, p21, and SA-β-Gal staining in liver sections or resected liver lobes (SA-β-gal wholemount staining) 6 days following delivery of NRASG12V and treated with control LNA (LNA-ctrl) or LNA targeting Snora24 (LNA-24). Graph shows mean ± SD mean intensity of SA-β-gal staining in liver from mice treated with LNA-ctrl (n = 3 mice) or LNA-24 (n = 3 mice) 6 days following NRASG12V expression. Statistical analysis was performed using an unpaired Student’s t-test, p=0.005.