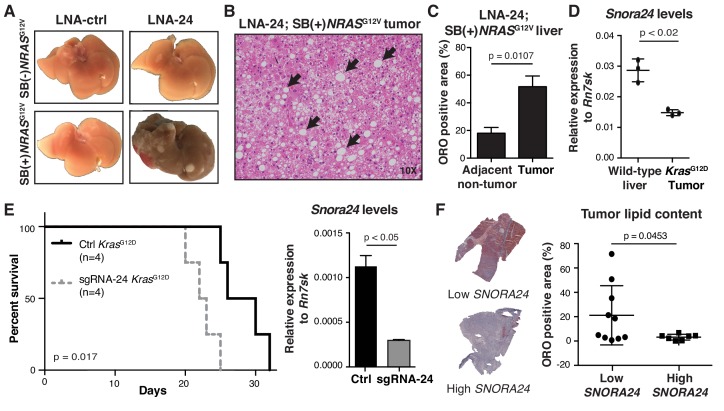
Figure 2. Snora24 plays a role in the initiation and maintenance of RAS-driven hepatocellular carcinoma.
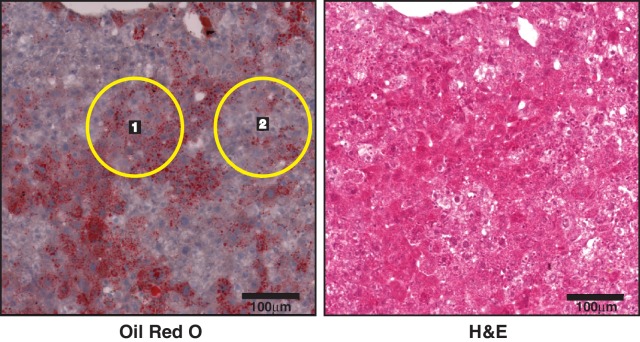
(A) Representative images of explanted livers from control (SB(-)NRASG12V) or SB(+)NRASG12V mice treated with either LNA-ctrl or LNA-24. (B) H and E staining of a liver section from SB(+)NRASG12V mouse treated with LNA-24. Black arrows highlight the presence of fat droplets. (C) Graph shows mean ± SD percentage Oil Red O (ORO) positive area in liver tumor nodules and adjacent non-tumor tissue from n = 3 SB(+)NRASG12V mice treated with LNA-24. For each mouse liver section, the amount of ORO positive stain per total area from at least four distinct tumor and non-tumor regions (as determined by H and E staining) was measured (see Materials and methods and Figure 2—figure supplement 1). Statistical analysis was performed using a paired Student’s t-test, p=0.0107). (D) Quantitative PCR (qPCR) analysis of Snora24 levels in wild-type liver or age- and sex-matched liver tumors from Alb-cre;KrasG12D mice. Graph shows mean ± SD Snora24 expression normalized to the levels of Rn7sk from n = 3 mice per condition. Statistical analysis was performed using an unpaired Student’s t-test, p<0.02. (E) Kaplan-Meier curves showing survival in male C57BL/6 wild-type mice following intrahepatic orthotopic injection of Ctrl KrasG12D HCC cells (black line, n = 4 mice) and sgRNA-24 KrasG12D HCC cells (gray dashed line, n = 4 mice), p=0.017, log-rank test (left panel). qPCR analysis of Snora24 in Ctrl KrasG12D and sgRNA-24 KrasG12D HCC cells (right panel). Graph shows mean relative expression ± SD normalized to the levels of Rn7sk from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, *p < 0.05. (F) Representative image of ORO staining in HCC from a patient with high SNORA24 (bottom) or low SNORA24 (top) expression. Quantification of ORO stain in tissue sections from HCC patients dichotomized into high or low by identifying samples with SNORA24 expression greater than ±one SD from the mean (n = 17 HCC specimens). Graph shows mean ± SD percentage Oil Red O (ORO) positive area present in HCC tissue specimens from patients with high or low SNORA24 expression and statistical analysis was performed using an unpaired Student’s t-test, p=0.0453.