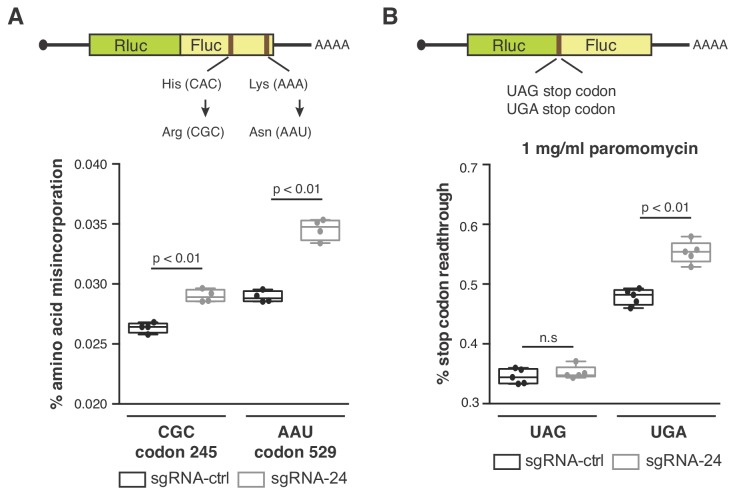
Figure 5. SNORA24-guided modifications impact translation accuracy.
(A) Diagram of luciferase reporters used to monitor amino acid misincorporation. Point mutations in Fluc at either His 245 or Lys 529 to near-cognate codons are highlighted (top panel). Amino acid misincorporation (%) of the indicated luciferase reporters (CGC codon or AAU codon) in HuH-7 sgRNA-ctrl cells (black) and HuH-7 sgRNA-24 cells (gray) are shown (bottom panel). On the box and whisker plots, the center line is the medium amino acid misincorporation (%), box limits are minimum and maximum values, and whiskers are S.D. from four independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p<0.01. (B) Diagram of luciferase reporters used to monitor stop codon readthrough (top panel). Stop codon readthrough (%) of the indicated luciferase reporters (UAG stop codon or UGA stop codon) in the presence of 1 mg/ml paromomycin in HuH-7 sgRNA-ctrl cells (black) and HuH-7 sgRNA-24 cells (gray) are shown (bottom panel). On the box and whisker plots, the center line is the medium stop codon readthrough (%), box limits are minimum and maximum values, and whiskers are S.D. from five independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p<0.01 and n.s = non significant.