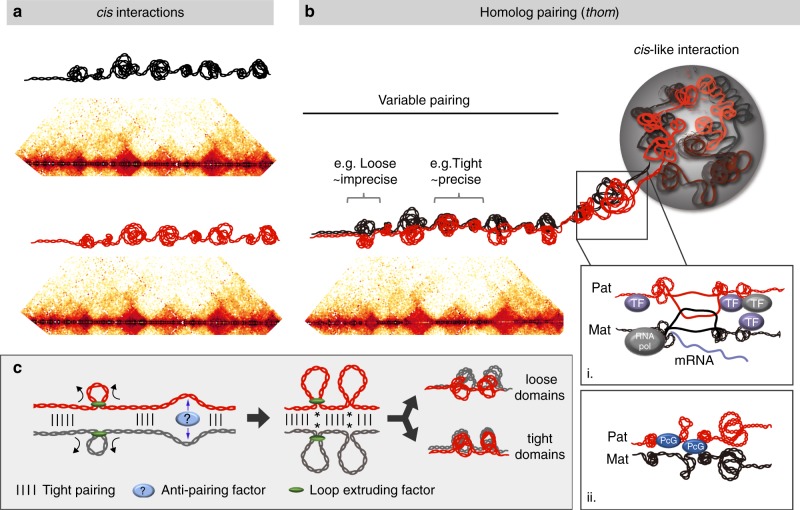
Fig. 4.
Haplotype-resolved Hi-C in PnM cells distinguished different forms of thom from cis interactions. a Cis contact maps for two homologs and schematics depicting possible cis interactions. b Thom contact map demonstrating a variable structure of pairing, including tight, precise pairing interspersed with looser, less precise pairing. Thom interactions encompass organizational structures that are concordant with cis behavior and could facilitate a variety of transcriptional states, including i. active or ii. repressive environments. c Left: Homologous loops between tightly paired regions may form by extrusion (black arrows), anti-pairing (blue arrows), or a combination of both. Right: Here, loops could result in thom domains that are either loosely paired (top), or railroad-track paired throughout if they fold back on each other, (bottom), with cis-maternal and cis-paternal domain boundaries concordant in both scenarios. Note, loop extrusion in mammalian systems is proposed to involve a cohesin ring79,80 (green oval) through which a single chromosome passes, suggesting that extrusion in the context of pairing may involve the passage of each homolog separately (shown) or two homologs simultaneously (not shown), the outcome of which could be either loosely or tightly paired domains. In a nonexclusive alternative, loops are formed by anti-pairing29, where pairing might be counterbalanced with unpairing via anti-pairing factors such as Cap-H2. Interestingly, extrusion and/or anti-pairing could bring enhancers and promoters together at the base of the loops (indicated by *), activating transcription, such as might happen in tightly paired regions. In the context of anti-pairing, this could explain the curious co-localization of Cap-H2 with regions of tight pairing (Supplementary Table 3; also ref. 27)