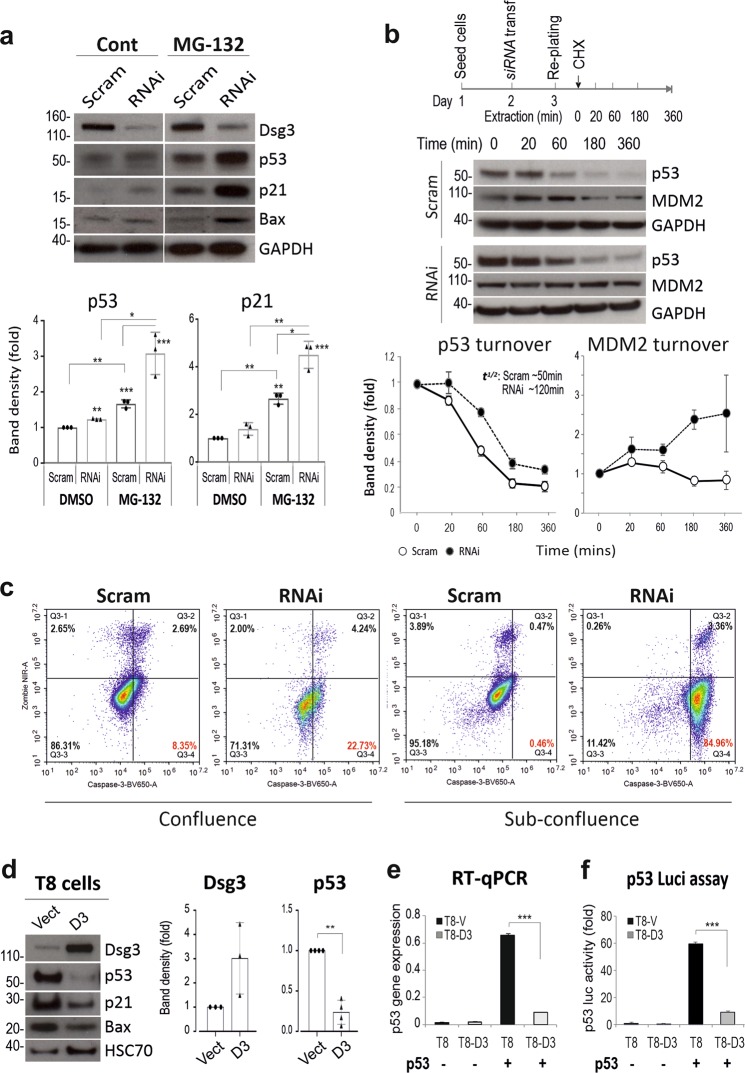
Fig. 2. The p53 suppression by Dsg3 was further supported by the inhibition of protein degradation and overexpression of Dsg3.
a Western blots of siRNA-transfected cells with and without treatment of MG-132 (25 µM) for 3 h. GAPDH and HSC70 were the loading controls. b Protein turnover analysis for p53, as well as MDM2, indicated reduced p53 turnover accompanied by MDM2 stabilization in Dsg3 depleted cells. The above is the timeline of the experiment. The band density for each blot was normalized against the loading control in each sample and then against the one at 0 min time point in each condition. The calculated half-life for p53 and MDM2 were shown in the graphs. c Flow cytometric analysis of cell viability with live cells (Zombie NIR−ve/Caspase-3−ve), Zombie NIR−ve/Caspase-3+ve, both positive (Zombie NIR+ve/Caspase-3+ve) and Zombie NIR+ve/Caspase-3−ve in NTERTs with and without Dsg3 knockdown, grown at 100% or ~40% confluences (the represented data of 3 independent attempts). d Protein expression in cutaneous keratinocytes T8 (p53 null, with p53 transfection) Vect control and Dsg3 overexpression (D3) that showed suppression of p53/p21WAF1/CIP1 in D3 cells compared to Vect cells. e RT-qPCR analysis of p53 expression (mean ± s.e.m.) in T8 cell lines (n = 3 independent assays of duplicate in each test). f p53 luciferase assay (mean ± s.d.) in T8 cell lines (n = 3, a representative of two independent experiments). The comparison was via unpaired two-sided student t-test. (**p < 0.01, **p < 0.01, ***p < 0.001)