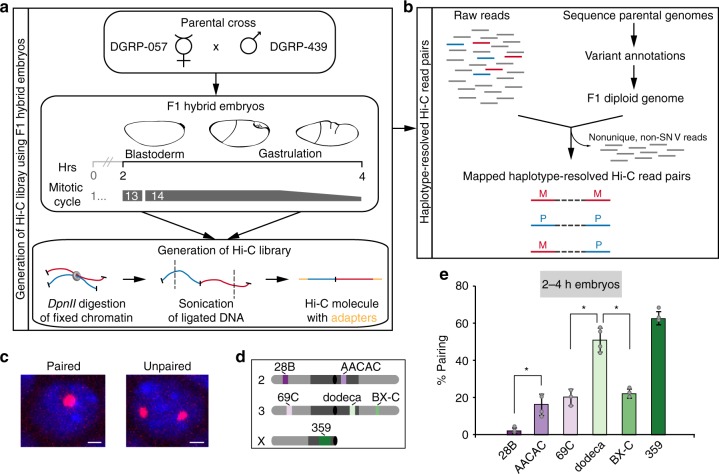
Fig. 1.
Haplotype-resolved Hi-C to characterize embryonic homolog pairing. a Generation of Hi-C libraries using 2–4 h hybrid embryos and b haplotype-resolved mapping. c Homolog pairing as assessed by FISH. Nuclei are considered paired nuclei when FISH signals are ≤0.8 μm (center-to-center distance) apart. Bar = 1 μm. d Location of FISH targets (heterochromatin, dark gray; euchromatin, light gray; centromere, black circle). e Percentage of nuclei showing paired loci in 2–4 h embryos (error bars, standard deviation of at least three replicates; n ≥ 100 nuclei/replicate; *P < 0.0001, Fisher’s two-tailed exact). Source data are provided as a Source Data file