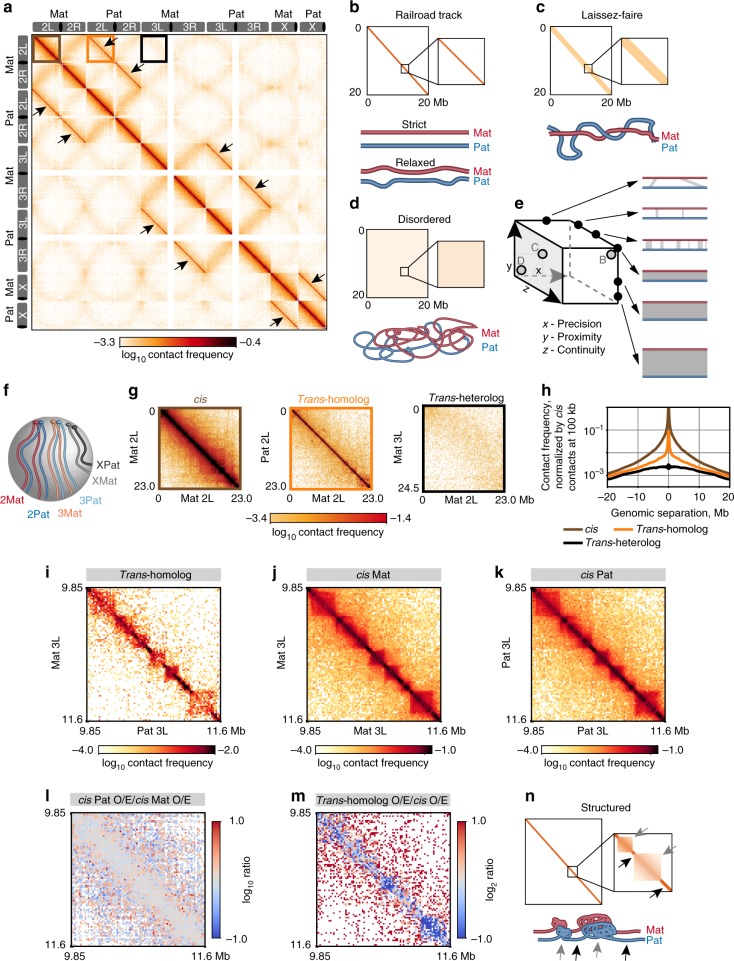
Fig. 3.
Highly structured homolog pairing resembles features of cis-organization. a Haplotype-resolved Hi-C map of F1 hybrid embryos. Arrows, trans-homolog diagonals. b–d Homologs juxtaposed in a b railroad track fashion, c a loose, laissez-faire mode, or d a highly disordered structure. e Homolog pairing may encompass a range of structures defined by precision, proximity, and continuity. f Rabl positioning of chromosomes. The g zoomed-in maps of chromosomal arms (color coded boxes in a; L, left; R, right) and h respective contact frequencies as a function of genomic distance (see Methods). i trans-homolog, j cis maternal, and k cis paternal maps of matching regions on chr3L. l Ratio of cis Pat/cis Mat Hi-C maps indicates the cis contact patterns of two homologs are highly consistent. m The ratio of trans-homolog/average cis maps suggests that pairing resembles cis contacts, albeit with lower interactions in some regions (dark blue). n Homolog pairing displays highly structured trans-domains (black) and trans-boundaries (gray), reflecting cis-organization of homologs