Fig. 1.
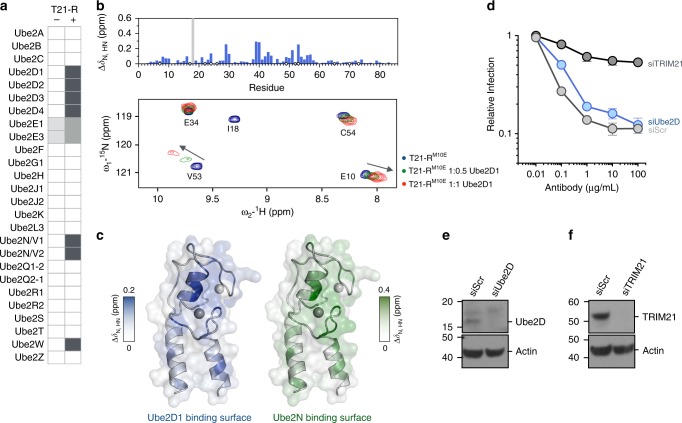
TRIM21 binds and catalyzes ubiquitination with physiologically redundant E2s. a In vitro E2 enzyme screen (no catalysis, white; catalysis, grey). Full blots are given in Supplementary Fig. 1. b A part of 15N-HSQC spectra of T21-RM10E in absence (blue) and presence of 0.5 (green) or 1 (red) molar equivalents of Ube2D1. Histogram of the chemical shift perturbations is shown against the primary structure. Blue circles indicate proline residues, white circles missing assignments. The gray bar in the histogram represents intermediate exchange of the amide of I18, as inferred from the disappearance of the I18 signal upon addition of Ube2D1. c The amide chemical shift perturbation upon E2 titration is mapped onto the T21-R structure in blue for the Ube2D1 titration (shown in Fig. 1b) and green for the Ube2N titration (shown in Supplementary Fig. 3). d Antibody (9C12)-dependent adenovirus 5 (Adv5) neutralization in HeLa cells stably transduced with small interfering RNA (siRNA) targeted against Ube2D or TRIM21. The data represent the mean ± s.e.m. from three independent experiments and normalized to virus only. Immunoblots of siRNA-mediated protein depletion of e Ube2D and f TRIM21. SiScr, scramble control siRNA. Source data are provided as a Source Data file