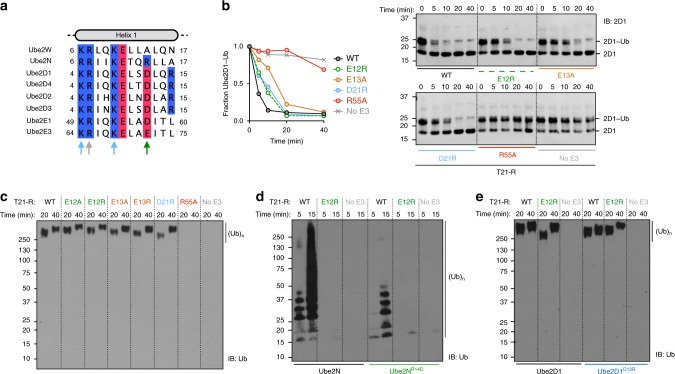
Fig. 5.
Ionic anchor points enable TRIM21 with a Ube2N-specific catalytic mechanism. a Clustal omega sequence alignment of all positive hits of the E2 screen (Fig. 1a). Arrows mark residues involved in RING interactions in our T21-R Ube2N~Ub structure. Lysine and arginine residues are colored in blue, aspartate and glutamate residues in red. The full alignment of all E2 enzymes used in the E2 screen (Fig. 1a) is shown in Supplementary Fig. 9. b Catalysis of ubiquitin discharge from pre-charged Ube2D1~Ub, and c catalysis of unanchored ubiquitin chains by Ube2D1 of T21-R anchor point mutants. Catalysis of ubiquitin chains by d Ube2N WT and R14D mutant and e Ube2D1 WT and D12R mutant. All biochemical assays were performed in at least two independent experiments. Source data are provided as a Source Data file