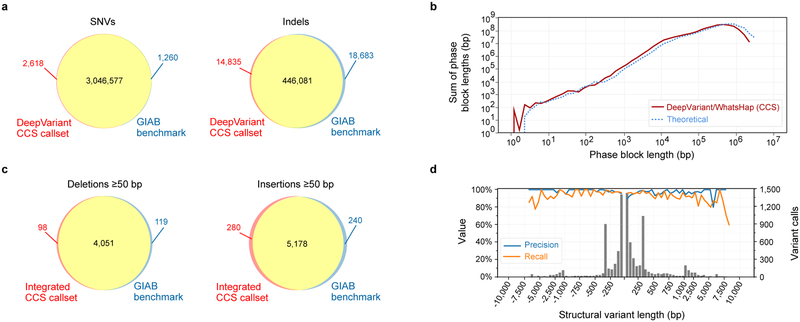
Figure 3. Variant calling and phasing with CCS reads.
(a) Agreement of DeepVariant SNV and indel calls with Genome in a Bottle v3.3.2 benchmark measured with hap.py. (b) Phasing of heterozygous DeepVariant variant calls with WhatsHap, compared to theoretical phasing of HG002 with 13.5 kb reads. (c) Agreement of integrated CCS structural variant calls with the Genome in a Bottle v0.6 structural variant benchmark measured with Truvari, (d) by variant length. Negative length indicates a deletion; positive length indicates an insertion. The histogram bin size is 50 bp for variants shorter than 1 kb, and 500 bp for variants >1 kb. All comparisons to GIAB are for the benchmark subset of the genome.