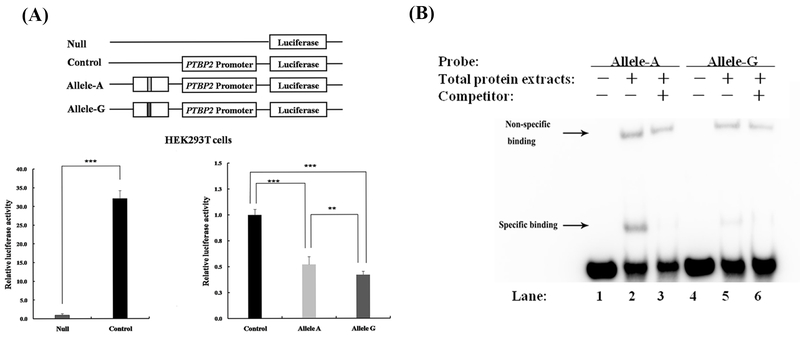
Figure 4. Dual-luciferase reporter assay and EMSA results.
(A) Dual-luciferase reporter assay result. The pGL3-PTBP2-Promoter vector was set as a control. Sequence construct containing either allele A or allele G was inserted upstream of the gene promoter. Eight replicates were conducted under each of the three conditions. Compared to the null pGL3-basic vector, the vector containing the PTBP2 promoter increases luciferase expression (p=3.65×10−7). Both allele A construct and allele G construct significantly reduce luciferase activity (both p<0.01), suggesting that the sequence containing rs12409479 represses PTBP2 promoter activity. Between the two allele constructs, the expression of allele A is significantly higher than that of allele G (p=4.15×10−3), implying that the regulation is allelic specific. (B) EMSA result. EMSA was conducted using a biotin labeled A-allele probe or G-allele probe with total protein extracted from HEK293T cells, with or without unlabeled A-allele probe or G-allele probe competitor. The shifted bands at the same migration position are observed in labeled probes for both allele A (lane 2) and allele G (lane 5) when compared to the controls (lane 1 and lane 4), implying their binding affinity to the same transcription factor. The two shifted bands are completely eliminated by the addition of 100-fold excess unlabeled A-allele probe (lane 3) and G-allele probe (lane 6), demonstrating that the binding is specific to the sequence motif containing rs12409479. Between the two alleles, the signal intensity for allele A (lane 2) is stronger than that for allele G (lane 5, too weak to be observed without zooming in the figure), suggesting that the former is more capable of binding to transcription factor than the latter. *: p<0.05; **: p<0.01; ***: p<0.001.