Figure 6. Rab35 interacts with Arl13b.
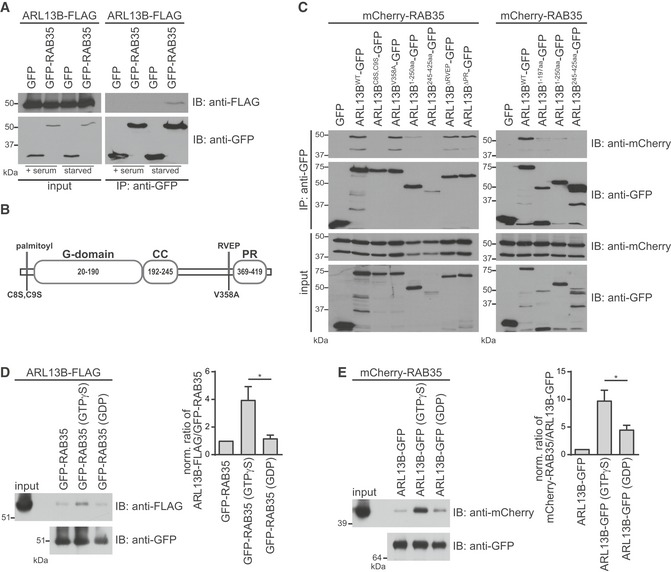
- HEK293T cells were transiently co‐transfected with ARL13B‐FLAG and GFP or GFP‐RAB35. Cells were cultured either in full growth medium (+serum) or serum‐starved for the final 8 h before harvesting. Immunoprecipitations (IP) were performed using anti‐GFP beads. Cell lysates (2.5% of input) and immunoprecipitates were loaded on the same gel, subjected to immunoblot (IB) analysis with indicated antibodies and detected with the same exposure.
- Schematic representation of the human ARL13B structure depicting the guanine nucleotide‐binding (G), coiled coil (CC) and proline‐rich (PR) domains as well as the point mutations affecting N‐terminal palmitoylation or C‐terminal ciliary‐targeting RVEP motif.
- HEK293T cells were transiently co‐transfected with mCherry‐RAB35 together with GFP or indicated ARL13B‐GFP wild‐type, mutant or truncation constructs. Cells were serum‐starved for the final 8 h before harvesting. IPs were performed using anti‐GFP beads and interacting proteins detected by immunoblotting (IB).
- HEK293T cells were transiently transfected with ARL13B‐FLAG and serum‐starved for the final 8 h before harvesting. HEK293T cell lysates expressing ARL13B‐FLAG were subjected to pull‐down with GFP‐RAB35 bound to anti‐GFP beads and preloaded with either no nucleotide, GTPγS or GDP. Bound proteins and cell lysates (1% of input) were analysed by immunoblotting. The graph shows the ratio of precipitated ARL13B and RAB35, normalised to the no nucleotide control. Data are mean ± SEM of three independent experiments. Statistical significance according ANOVA followed by Tukey post hoc test (*P = 0.0333).
- HEK293T cells were transiently transfected with mCherry‐RAB35 and serum‐starved for the final 8 h before harvesting. HEK293T cell lysates expressing ARL13B‐FLAG were subjected to pull‐down with ARL13B‐GFP bound to anti‐GFP beads and preloaded with either no nucleotide, GTPγS or GDP. Bound proteins and cell lysates (1% of input) were analysed by immunoblotting. The graph shows the ratio of precipitated ARL13B and RAB35, normalised to the no nucleotide control. Data are mean ± SEM of three independent experiments. Statistical significance according ANOVA followed by Tukey post hoc test (*P = 0.0466).
Source data are available online for this figure.
