Figure 2. Absence of PCGF3/5‐PRC1 proteins from the Xist ΔB+C RNA protein interactome.
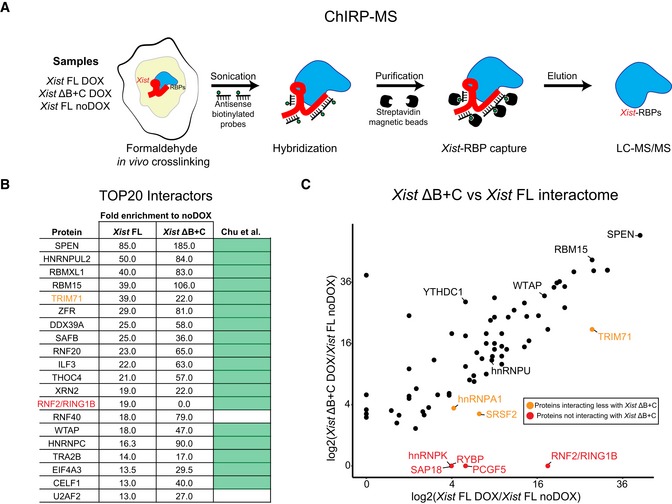
- Scheme of the ChIRP‐MS workflow performed on Xist FL (DOX and noDOX conditions) and Xist ΔB+C (DOX) at day 3 of differentiation; RBP—RNA‐binding protein.
- Top 20 protein hits from the ChIRP‐MS of Xist FL; the ranking was based on fold enrichment of Xist FL DOX versus Xist FL noDOX; weakly annotated protein isoforms with an Annotation score in UniProtKB < 3 (out of 5) were excluded; fold enrichment for Xist ΔB+C is also displayed for comparison; light green boxes correspond to proteins previously described by Chu et al 13 as Xist interactors; protein in red (RNF2/RING1B) represents a protein not found in the Xist ΔB+C interactome; protein in light brown (TRIM71) is less enriched in Xist ΔB+C than in Xist FL.
- Scatter plot displaying the differences in peptide counts between Xist FL and Xist ΔB+C for the 74 out of 81 Xist interactors from Chu et al 13 with a minimum of fold change of 2.5 in Xist FL or Xist ΔB+C; shown is the log2 fold change in peptide counts of each mutant in DOX conditions compared with the Xist FL in noDOX conditions; proteins retrieved by both Xist FL and Xist ΔB+C ChIRPs with a proposed role in XCI such as SPEN, RBM15, WTAP, YTHDC1, and hnRNPU are indicated; light brown dots mark proteins more represented in Xist FL than in Xist ΔB+C ChIRPs, while red dots display proteins which are only retrieved by Xist FL ChIRP.
