Figure 3. Strong hydrophobic interactions are contributing specificity and high affinity between the CENP‐A nucleosome and CENP‐CCR .
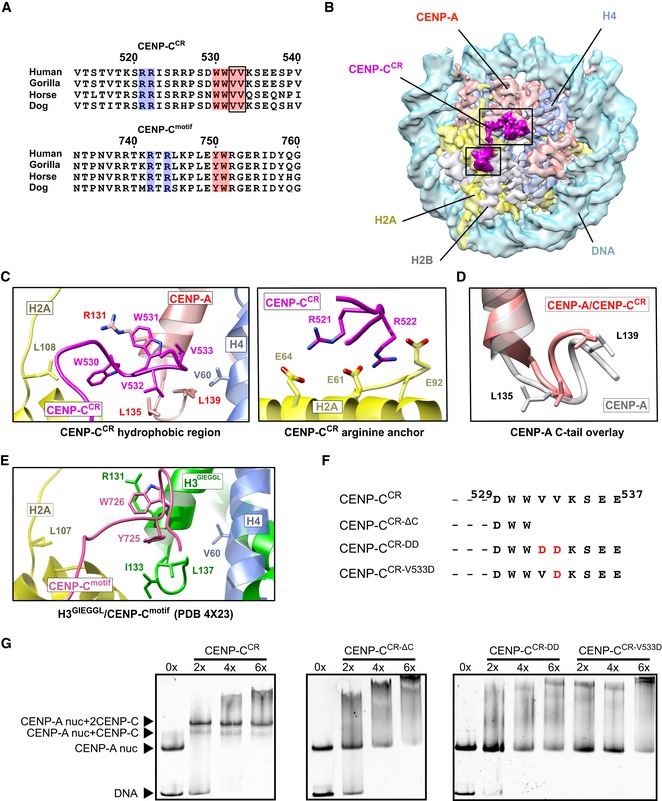
- Sequence alignment of CENP‐CCR and CENP‐Cmotif regions from different mammals. Conserved residues involved in CENP‐A binding are highlighted in blue (electrostatic interactions) and pink (hydrophobic interactions). Residues identified in this study to contribute higher affinity of CENP‐CCR comparing to CENP‐Cmotif are boxed.
- Cryo‐EM density map of the human CENP‐A nucleosome, color‐coded for histones, in complex with CENP‐CCR (magenta). CENP‐CCR binds CENP‐A nucleosome through hydrophobic region (big box) and arginine anchor (small box). Interacting residues in each of the regions are shown in (C).
- Ribbon diagram showing interactions of the CENP‐CCR hydrophobic region (left) and CENP‐CCR arginine anchor (right) with the CENP‐A nucleosome.
- Overlay of the CENP‐A C‐terminal tail before (gray) and after (red) binding of CENP‐CCR.
- Interactions between H3‐GIEGGL and rat CENP‐Cmotif as observed in PDB 4X23 14.
- Schematic diagram of mutated CENP‐CCR sequences used to test importance of CENP‐CV532 and CENP‐CV533 for generation of CENP‐A/CENP‐CCR complexes.
- Native gels showing the impaired ability of mutated CENP‐CCR to form complexes with the CENP‐A nucleosome. The molar ratio of CENP‐CCR/CENP‐A nucleosomes is shown above each lane.
