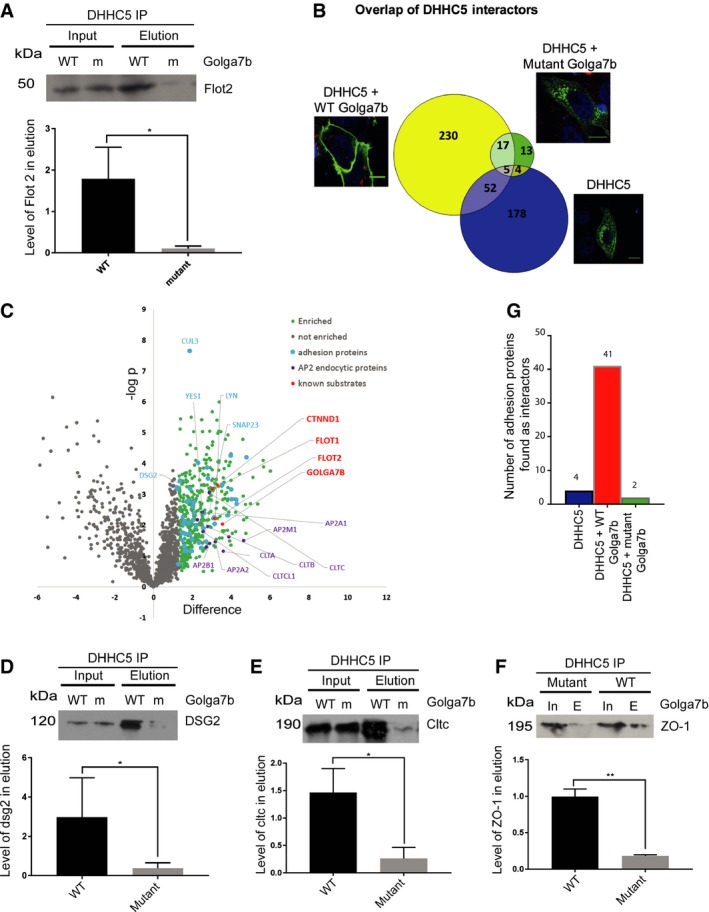
FLAG‐DHHC5 immunoprecipitations were performed using lysates from Hela cells co‐expressing either wild‐type (WT) or palmitoylation‐deficient mutant (m) Golga7b. The interaction of DHHC5 with its substrate Flot2 was measured by immunoblotting and is significantly reduced in the presence of palmitoylation‐deficient mutant Golga7b, quantification normalised to input, *P < 0.05, ratio paired t‐test, n = 3.
Venn diagram comparing the overlap of numbers of proteins immunoprecipitated when FLAG‐DHHC5 is expressed alone or co‐expressed with either WT or mutant Golga7b. IF images are taken from Figs
EV2 and
EV3 to illustrate DHHC5 localisation.
Volcano plot of DHHC5 interactors from AP‐MS analysis from HeLa cells co‐expressing FLAG‐DHHC5 and WT Golga7b highlighting interacting proteins involved in clathrin‐mediated endocytosis, cell:cell adhesion and known substrates of DHHC5.
Validation of novel DHHC5 interactors by co‐IP: immunoblots of Dsg2 from Hela cells co‐expressing DHHC5 and either wild‐type (WT) or mutant (m) Golga7b and quantification normalised to input, *P < 0.05, ratio paired t‐test, n = 3.
Co‐IP immunoblots of clathrin heavy chain from Hela cells co‐expressing DHHC5 and either wild‐type (WT) or mutant (m) Golga7b and quantification normalised to input, *P < 0.05, ratio paired t‐test, n = 3.
Co‐IP immunoblots of ZO‐1 protein from Hela cells co‐expressing DHHC5 and either wild‐type (WT) or mutant (m) Golga7b and quantification normalised to input, **P < 0.01 ratio paired t‐test, n = 3.
Bar graph comparing the number of the cell:cell adhesion proteins identified in each protein interaction dataset.
Data information: All scale bars: 10 μm. All error bars represent SEM.