-
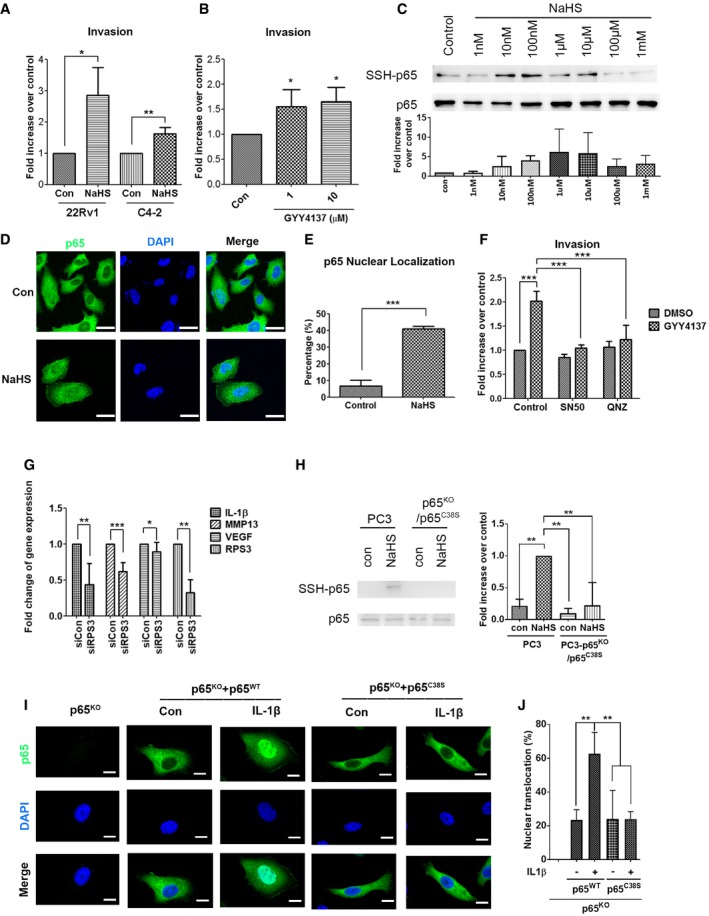
A
22Rv1 and C4‐2 cells were incubated for invasion assay for 16–18 h. DMEM/10% FBS, together with 1 μM NaHS served as a chemoattractant.
-
B
PC3 cells were incubated for invasion assay for 24 h. DMEM/10% FBS, together with 1 or 10 μM GYY4137, served as a chemoattractant.
-
C
Top: PC3 cells treated with 1 nM–1 mM NaHS for 1 h were subjected to the modified biotin switch assay with the antibody against p65 to detect S‐sulfhydration. Bottom: Quantitative analysis of SSH‐p65 protein level, and normalized with total p65 level.
-
D
PC3 cells were treated with 1 μM NaHS for 24 h in the absence of serum. Subcellular localization of p65 was detected by immunocytochemistry. Nuclei were counterstained with DAPI. Scale bar: 25 μm.
-
E
Nuclear translocation of p65 was scored by counting the number of nuclear positive stained cells of p65 to the total number of cells in random microscopic fields.
-
F
PC3 cells were incubated for invasion assay for 24 h. DMEM/10% FBS, together with 1 μM GYY4137 and NF‐κB inhibitors (50 μg/ml SN50, or 100 nM QNZ), served as a chemoattractant.
-
G
Real‐time RT–PCR analysis for IL‐1β, MMP‐13, VEGF, and RPS3 mRNA level in PC3 cells with control or RPS3 knockdown.
-
H
Left: PC3 cells with p65 knockout with p65 C38S mutant and PC3 parental cells were treated with 100 μM NaHS for 1 h and subjected to the modified biotin switch assay with the antibody against p65 to detect S‐sulfhydration. Right: Quantitative analysis of SSH‐p65 protein level, and normalized with total p65 level.
-
I
PC3 cells with p65 knockout were transfected with vector, p65 wild‐type, or p65 C38S mutant and then exposed to IL‐1β (20 ng/ml) for 1 h. Subcellular localization of p65 was detected by immunocytochemistry. Nuclei were counterstained with DAPI. The representative images are shown. Scale bars: 10 μm.
-
J
Nuclear translocation of p65 was scored by counting the nuclear positive stained cells and the total number of cells quantified in random microscopic fields.
= 3 biological replicates). Student's
‐test (A, E, G) or ANOVA followed by Tukey's post hoc test (B, F, H, I) was used for statistical analysis (*
< 0.001).