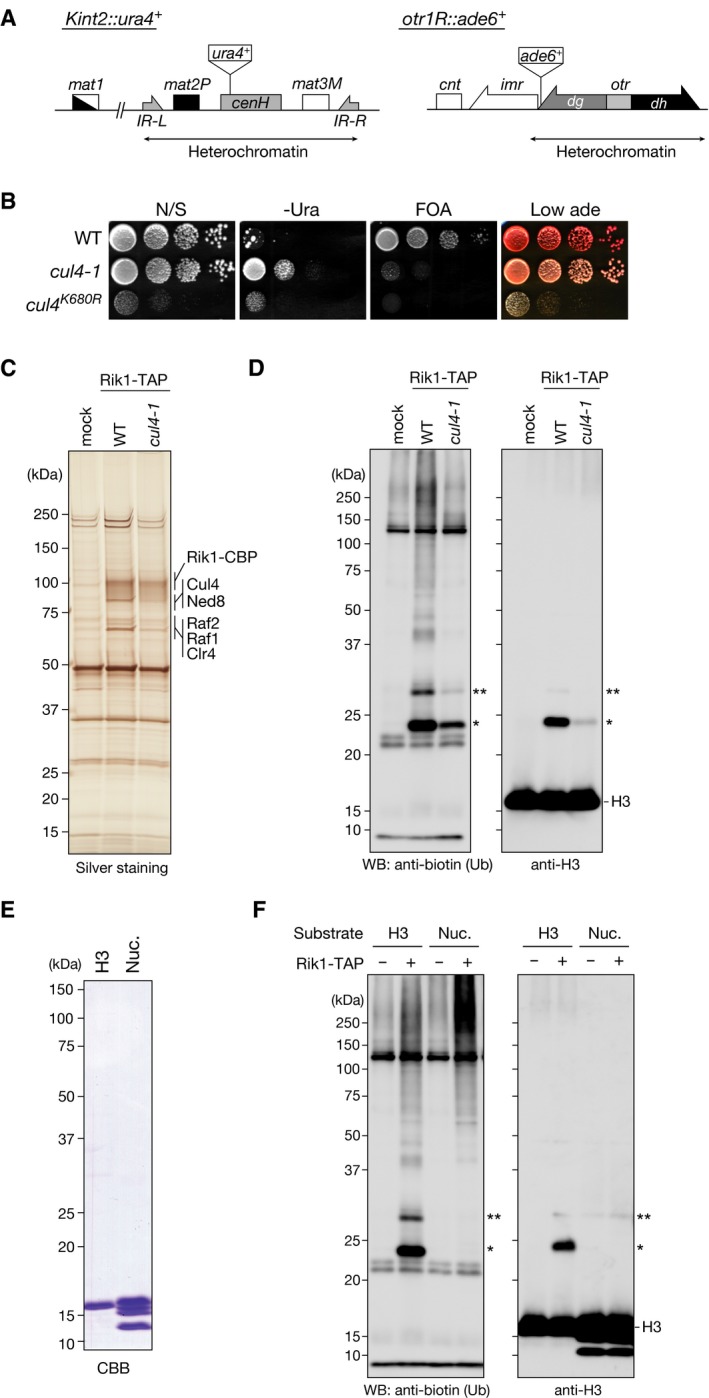
Diagram of the mating‐type loci and part of centromere 1 in S. pombe. The positions of the silencing reporter genes (Kint2::ura4
+ and otr1R::ade6
+) are shown.
Heterochromatic silencing assays of wild‐type and cul4 mutant strains. Silencing at the mating‐type Kint2::ura4
+ and otr1R::ade6
+ was evaluated. Ten‐fold serial dilutions of the indicated strains were spotted onto non‐selective medium (N/S), medium lacking uracil (‐Ura), medium containing 5‐FOA (5‐FOA), and medium containing low adenine (Low ade).
Purified TAP‐tagged Rik1‐containing complexes analyzed by SDS–PAGE and silver staining. mock: a mock purification from an untagged strain. Proteins identified by LC‐MS/MS are indicated at right.
In vitro ubiquitylation assays using biotinylated ubiquitin, purified CLRC, and recombinant S. pombe histone H3 as the substrate. Proteins were analyzed by Western blotting using the indicated antibodies. The single and double asterisks indicate mono‐ and di‐ubiquitylated histone H3 species, respectively.
Recombinant histone H3 protein and core histones in the reconstituted nucleosomes (Nuc.) were analyzed by SDS–PAGE and visualized by CBB staining.
In vitro ubiquitylation assays using biotinylated ubiquitin, purified CLRC, and recombinant S. pombe histone H3 or reconstituted nucleosomes (Nuc.) as the substrate. Proteins were analyzed by Western blotting using the indicated antibodies. The single and double asterisks indicate mono‐ and di‐ubiquitylated histone H3 species, respectively.