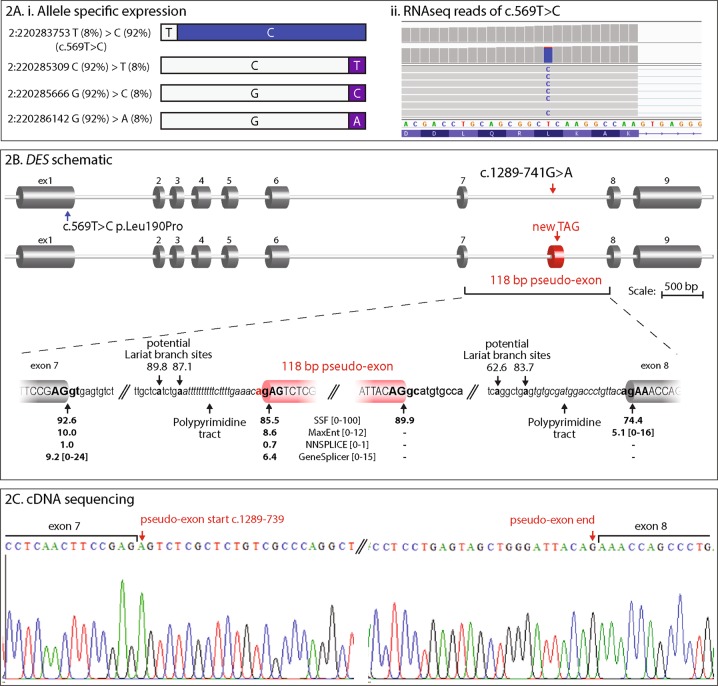
Fig. 2.
a Disparate patterns of mRNA expression levels from DES alleles. (i) Schematic representation of allelic percentages derived from RNA-seq read depth. The paternal g.220283753 T > C, c.569 T > C variant shows allele bias, accounting for 92% of RNA-seq reads. Three other common SNPs in the coding region also show allele bias, with the reference sequence present in 92% of reads. Inheritance of these SNPs could not be determined; although allele bias data infers maternal inheritance. No splicing aberrations were observed to result from these common SNPs. (ii) Screenshot showing the RNA sequencing reads over the paternal missense c.569 T > C variant and substantial allele bias of variant C nucleotide. b Schematic representation of DES exon structure and consequence of the DES intronic variant c.1289-741 G > A, r.1288_1289ins1289-739_1289-622 (118 bp artificial exon/intronic fragment and abnormal termination codon). Enlarged representation of the artificial exon/intronic fragment and flanking essential splice sites. Predicted strengths of the splice donor and accepter using Alamut Visual® Interactive Biosoftware are annotated beneath. C: cDNA sequencing electropherogram of a RT-PCR amplicon showing inclusion of the 118 nt artificial exon/intronic fragment into the DES mRNA, flanked by exon 7 and exon 8 sequences