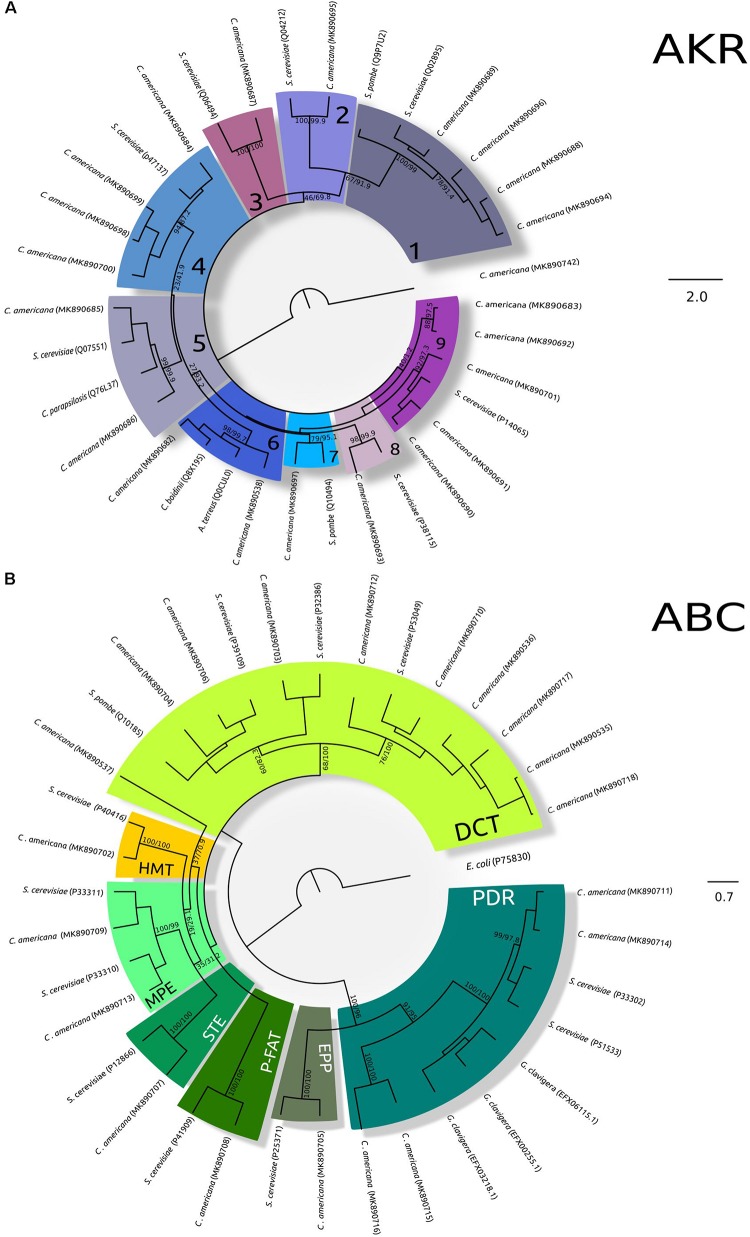
FIGURE 2.
Maximum-likelihood trees of AKR and ABC transporters. (A) Phylogeny of AKR. The analysis was performed using the amino acid substitution model LG + G + F with a gamma parameter of 1.582, bootstrap/aLRT SH-like values shown at nodes. The alcohol dehydrogenase (non-member of the AKR Superfamily) (from C. americana was used as outgroup. 1, Aryl-alcohol dehydrogenase; 2, D-arabinose 1-dehydrogenase; 3, Pyridoxal reductase; 4, Oxidoreductase; 5, NAD(P)H-dependent reductase; 6, NAD(P)H-dependent D-xylose reductase; 7, Aldehyde reductase I; 8, D-arabinose dehydrogenase (NAD(P)+) heavy chain; 9, Glycerol 2-dehydrogenase (NAD(P)+). Accession numbers of TCDB/GenBank sequences are shown in brackets. (B) Phylogeny of ABC transporters. The analysis was performed using the amino acid substitution model VT + G + I + F with a gamma parameter of 2.312 and a proportion of invariable sites of 0.005, bootsrap/aLRT SH-like values shown at nodes. The ABC III from E. coli (P75830) was used as outgroup. DCT, drug conjugate transporter; P-FAT, peroxisomal fatty acyl CoA transporter; HMT, heavy metal transporter; MPE, mitochondrial peptide exporter; STE, a-factor sex pheromone exporter; PDR, pleiotropic drug resistance; EPP, eye pigment precursor transporter. Accession numbers of TCDB/GenBank sequences are shown in brackets.)