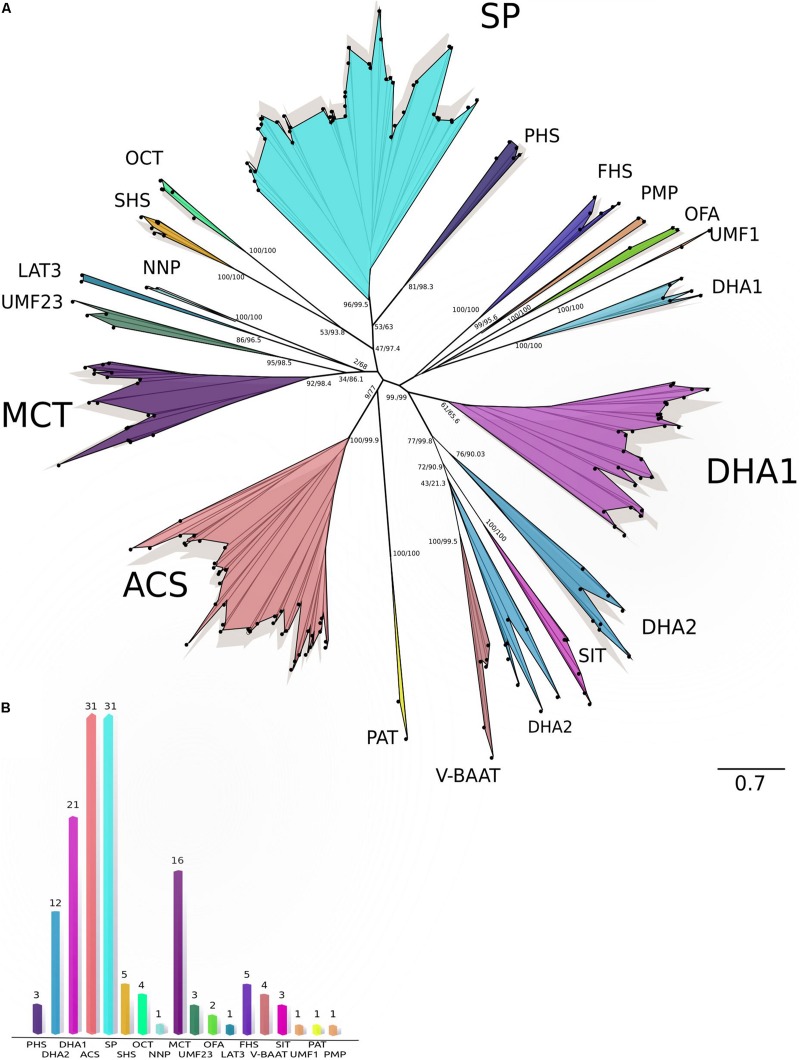
FIGURE 3.
Maximum-likelihood tree and Classification of MFS transporters. (A) Phylogeny of MFS sequences. The maximum-likelihood tree of MFS based on amino acid sequences from C. americana plus TCDB sequences. The analysis was performed using the amino acid substitution model VT + G + F with a gamma (parameter of 3.876. Bootstrap/aLRT SH-like values are shown at nodes. (B) Classification of MFS families. The graph shows 145 putative MFS classified into 18 different MFS families. ACS, anion:cation symporter family; DHA1 and DHA2, drug:H+ antiporter 1–2 families; FHS, fucose:H+ symporter family; LAT3, L-amino acid transporter-3 family; MCT, monocarboxylate transporter family; NNP, nitrate/nitrite porter family; OCT, organic cation transporter family; OFA, oxalate:formate antiporter family; PAT, peptide-acetyl-coenzyme a transporter family; PHS, phosphate:H+ symporter family; PMP, putative magnetosome permease family; SHS, sialate:H+ symporter family; SIT, siderophore-iron transporter family; SP, sugar porter family; UMF1, unidentified major facilitator-1 family; UMF23, unidentified major facilitator-23 family; V-BAAT, vacuolar basic amino acid transporter family.)