Abstract
Bladder cancer (BCa) is a common urinary tract malignancy with frequent recurrences after initial resection. Submucosal injection of gemcitabine prior to transurethral resection of bladder tumor (TURBT) may prevent recurrence of urothelial cancer. However, the underlying mechanism remains unknown. In the present study, ultra-performance liquid chromatography Q-Exactive mass spectrometry was used to profile tissue metabolites from 12 BCa patients. The 48 samples included pre- and post-gemcitabine treatment BCa tissues, as well as adjacent normal tissues. Principal component analysis (PCA) revealed that the metabolic profiles of pre-gemcitabine BCa tissues differed significantly from those of pre-gemcitabine normal tissues. A total of 34 significantly altered metabolites were further analyzed. Pathway analysis using MetaboAnalyst identified three metabolic pathways closely associated with BCa, including glutathione, purine and thiamine metabolism, while gluta-thione metabolism was also identified by the enrichment analysis using MetaboAnalyst. In search of the possible targets of gemcitabine, metabolite profiles were compared between the pre-gemcitabine normal and post-gemcitabine BCa tissues. Among the 34 metabolites associated with BCa, the levels of bilirubin and retinal recovered in BCa tissues treated with gemcitabine. When comparing normal bladder tissues with and without gemcitabine treatment, among the 34 metabolites associated with BCa, it was observed that histamine change may be associated with the prevention of relapse, whereas thiamine change may be involved in possible side effects. Therefore, by employing a hypothesis-free tissue-based metabolomics study, the present study investigated the metabolic signatures of BCa and found that bilirubin and retinal may be involved in the mechanism underlying the biomolecular action of submucosal injection of gemcitabine in urothelial BCa.
Key words: tissue metabolomics, submucosal injection, gemcitabine, biomarkers, bladder cancer
Introduction
Bladder cancer (BCa) ranks ninth among the most common solid tumors worldwide (1). Approximately 75% of newly diagnosed BCa cases are non-muscle invasive, and the majority are histologically low-grade cancer (2). Routine surveillance to monitor BCa recurrence includes cystoscopic examination due to a high risk of recurrence after initial resection. The repeat transurethral resection of bladder tumor (TURBT) remains the first-line treatment for BCa recurrence (3). However, all these invasive procedures result in a high cost of care, and are often associated with significant morbidity. Therefore, more effective interventions to prevent BCa recurrence are urgently needed.
Submucosal injection of antitumor drugs (pirarubicin) after standard TURBT was proven to be an effective approach to reducing superficial tumor recurrence (4). Gemcitabine is a pivotal chemotherapeutic agent widely used for BCa due to its low toxicity in general (5) and as an intravesical instillation (3). Data from our experimental and clinical studies also demonstrated that submucosal injection of gemcitabine prior to TURBT significantly reduced BCa recurrence (6). However, the underlying mechanisms are largely unknown.
Metabolomics is a newly emerging technology, which enables the identification of endogenous compounds and potentially novel mechanisms associated with disease processes (7). Metabolomics has been used to profile metabolites in various biological samples, such as serum (8), urine (9-15) and tissue (16,17), which are the results of the metabolic response of living systems to drug toxicity or disease (10). Potential biomarkers identified from metabo-lomic profiling studies on BCa may be of diagnostic value and act as indicators of cancer recurrence (18). Currently, a number of analytical platforms, such as high-performance liquid chromatography/mass spectrometry (HPLC/MS) (10), ultra-performance LC-MS (UPLC-MS) (12), and UPLC time-of-flight MS (UPLC-TOF-MS) (15), have been employed to study the metabolomics of BCa by using urine samples. However, metabolomic studies on BCa tissues is relatively scarce (16,17). Notably, different metabolomic platforms with their unique analytical approaches provide complementary insights into metabolome changes (9,11-15). Therefore, there is a need to study the tissue-based metabolic signatures of BCa using a new metabolomics platform.
Metabolomics has also been applied to cancer treatment and drug target discovery. Eidelman et al reported using metabolomics to screen the potential therapeutic pathways in prostate cancer (PCa) (19). Metabolomics has also been proven to be a promising approach to developing reliable therapeutic targets for PCa treatment (20). The present study employed liquid chromatography (LC)-Q-Exactive MS-based metabo-lomic technology to study the metabolic changes in BCa tissues before and after treatment with gemcitabine. Identification of the key metabolites may reveal new metabolic changes associated with BCa and uncover the changes that mediate the effect of gemcitabine in the treatment of BCa.
Materials and methods
Clinical samples
A total of 12 patients (9 men and 3 women; age range, 55-85 years) who had undergone TURBT at the Affiliated Huai'an No. 1 People's Hospital of Nanjing Medical University were recruited between December 2016 and September 2017. Bladder tissue samples were collected from the same patient immediately prior to and 30 min after submucosal injection of gemcitabine (50 mg, dissolved in 20 ml normal saline). The bladder tissues included BCa as well as adjacent non-cancerous bladder tissues. Therefore, a total of 48 samples were collected in four groups: Pre-gemcitabine normal, pre-gemcitabine BCa, post-gemcitabine normal, and post-gemcitabine BCa. Histopathological diagnosis was conducted by two independent pathologists according to the classification criteria of the World Health Organization/International Society of Urological Pathology (21). Written informed consent was obtained from each participant prior to recruitment. The Ethics Committee of The Affiliated Huai'an No. 1 People's Hospital of Nanjing Medical University reviewed and approved the study protocol (serial no. YL-P-2013-21-01). The sample size of the present study was similar to that of a previous study on PCa tissues, which produced valuable findings (22). In addition, the experimental design of self-control and complete collection of samples in our study may improve statistical power by avoiding confounding factors.
Tissue preparation for metabolomic analysis
Following harvesting, all tissues were snap-frozen in liquid nitrogen, and kept in a -80°C freezer until further analysis. The sample preparation was conducted as described previously (23). Briefly, the tissues were fragmented, ultrasonicated for 5 min (power: 60%, pulses: 6/4) in distilled water, and then 150 μl homogenate and 450 μl methanol (Merck KGaA) were mixed in a 1.5-ml Eppendorf tube for protein precipitation. The mixture was centrifuged at 16,000 x g for 15 min at 4°C, and the supernatant was collected and dried in a vacuum centrifugal concentrator. The dry residue was reconstituted in ultra-pure water and used for metabolomic analysis.
Metabolomic analysis
The metabolomic analysis was conducted as previously reported (23). LC-HRMS analysis was performed on an UPLC Ultimate 3000 system (Dionex), coupled with a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Inc.). The instrument operated at a 70,000 resolution with a full-scan acquisition ranging from 70 to 1,500 m/z. The chromatographic separation of metabolites associated with the metabolomic profiling used a multistep gradient containing ultra-pure water (mobile phase A) and acetonitrile (ACN; mobile phase B), both acidified with 0.1% formic acid. The gradient operated at a flow rate of 0.4 ml/min over a 15-min period. The metabolites were identified based on the accurate mass and the retention time compared with the commercial standards. The metabolite standards were purchased from Sigma-Aldrich; Merck KGaA, Damas-beta Co., Ltd., Aladdin Reagent Company and Adamas Reagent Co., Ltd.
Statistical analysis
Data collected from the mass spectrometer were processed for pattern recognition analysis (principal component analysis, PCA). Normalized MS data were exported to SIMCA-P+ software (V14.0, Umetrics AB) to perform PCA where grouping trends could be observed. The difference in metabolites between two groups was compared by paired t-test. According to previous reports (24,25), the correlation between metabolite changes and cancer stage was analyzed based on the comparison between two groups using paired t-test based on the cancer stage classification. A P-value of <0.05 was considered as the threshold for statistically significant differences.
To further characterize the metabolic changes and the metabolic pathways involved, the differentiated metabolites were first annotated with Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) and Human Metabolome Database (HMDB, http://www.hmdb.ca/) (date of access for databases: April 19, 2018). Data were then processed and analyzed using MetaboAnalyst 4.0 (http://www.metaboanalyst.ca/MetaboAnalyst/) by R software (v3.4.3, GitHub). Two modules of MetaboAnalyst were used, namely pathway analysis and enrichment analysis, which are based on the KEGG database and Small Molecule Pathway Database (SMPDB, http://smpdb.ca/), respectively (26). The metabolic network of the differential metabolites and altered metabolic pathways in KEGG general metabolic pathway was visualized by iPath 3.0 (http://pathways.embl.de/).
Results
Clinical characteristics of 12 subjects
The mean age of the subjects, including 9 men and 3 women, was 67 years (range, 55-85 years). In all 12 patients, the diagnosis of urothelial carcinoma was confirmed by histopathological examination. A total of 2 patients had Ta, 1 patient had T1, and 9 patients had T2 disease. Two patients were diagnosed with high-grade T2 tumors with squamous metaplasia. The clinical characteristics of the participants are shown in Table I, and are considered to be representative according to the general population of BCa in China (2,27).
Table I.
Clinical characteristics of 12 BCa patients.
| Number | Sex | Age (years) | Tumor type | Histopathology | |
|---|---|---|---|---|---|
| Stage | Grade | ||||
| 01 | Male | 55 | MIUC | T2 | High |
| 02 | Male | 56 | MIUC | T2 | High |
| 03 | Male | 59 | MIUC | T2 | High |
| 04 | Female | 82 | MIUC | T1 | High |
| 05 | Male | 67 | MIUC | T2 | High |
| 06 | Male | 76 | MIUC | T2b | Squamous metaplasia |
| 07 | Male | 76 | MIUC | T2 | High |
| 08 | Female | 60 | MIUC | T2 | High |
| 09 | Male | 67 | NMIUC | Ta | High |
| 10 | Male | 58 | NMIUC | Ta | High |
| 11 | Male | 85 | MIUC | T2a | High |
| 12 | Female | 61 | MIUC | T2 | Squamous metaplasia |
MIUC, muscle-invasive urothelial carcinoma; NMIUC, non-muscle-invasive urothelial carcinoma. BCa, bladder cancer.
PCA
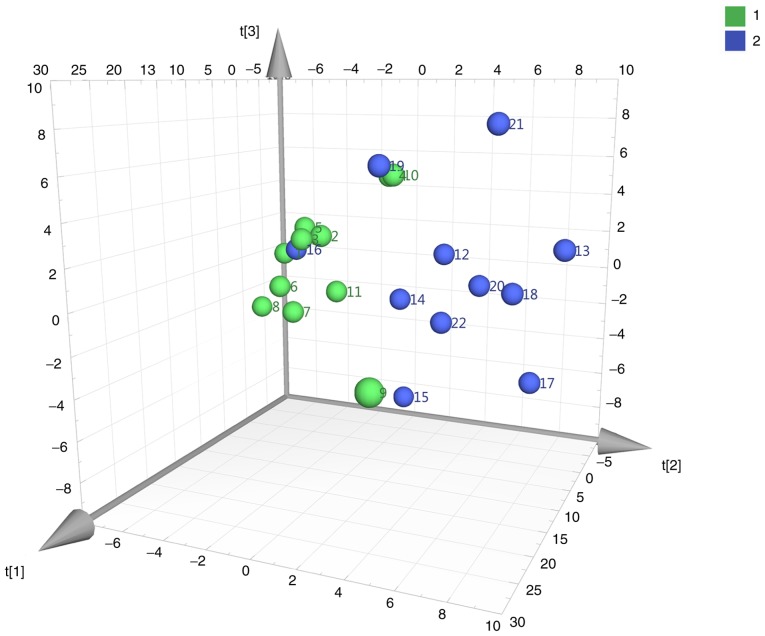
A total of 165 metabolites were detected. PCA was performed to process the metabolite data based on a mean center-scaling model, which is an unsupervised projection method employed to visually display the intrinsic similarities and differences in the dataset. As shown in Fig. 1, PCA (pre-gemcitabine normal vs. pre-gemcitabine BCa tissues) revealed a well-differentiated and clustered pattern in score plots, indicating the significant metabolome changes between these two groups.
Figure 1.
PCA score plots derived from pre-gemcitabine normal (green) and pre-gemcitabine BCa (blue) tissues. Comparison of pre-gemcitabine normal (green) and pre-gemcitabine BCa (blue) tissues. PCA, principal component analysis; BCa, bladder cancer.
Altered metabolites and cancer-associated metabolic pathways in BCa
UPLC-Q-Exactive analysis identified 34 differentially expressed metabolites annotated in the KEGG and HMDB databases in pre-gemcitabine BCa tissues compared with pre-gemcitabine adjacent normal tissues (Table II).
Table II.
List of the altered metabolites identified in BCa and their changes in the comparison between BCa tissues with or without gemcitabine pretreatment and pre-gemcitabine normal tissues.
| Metabolites | KEGG | HMDB | Pre-gemcitabine BCa vs. normal tissuesa | Post-gemcitabine BCa vs. pre-gemcitabine normal tissuesa | ||
|---|---|---|---|---|---|---|
| Fold changea | P-value | Fold changea | P-value | |||
| Deoxycytidine | C00881 | HMDB0000014 | 4.03 | 1.22E-02 | 10.10 | 4.15E-05 |
| 5′-Methylthioadenosine | C00170 | HMDB0001173 | 15.16 | 2.38E-04 | 12.22 | 4.16E-04 |
| 3′-AMP | C01367 | HMDB0003540 | 4.95 | 3.02E-02 | 6.36 | 3.33E-02 |
| Androstenedione | C00280 | HMDB0000053 | 0.16 | 4.09E-02 | 0.10 | 5.76E-03 |
| Bilirubin | C00486 | HMDB0000054 | 0.4 | 3.97E-02 | 1.26 | 2.04E-01 |
| Cholic acid | C00695 | HMDB0000619 | 0.29 | 3.14E-02 | 0.24 | 2.00E-03 |
| Cytidine | C00475 | HMDB0000089 | 3.83 | 2.43E-03 | 3.05 | 6.77E-03 |
| 5-Hydroxylysine | C16741 | HMDB0000450 | 0.64 | 4.40E-02 | 0.48 | 7.49E-03 |
| Deoxyinosine | C05512 | HMDB0000071 | 2.44 | 3.87E-02 | 4.12 | 1.70E-02 |
| Glucosamine 6-phosphate | C00352 | HMDB0001254 | 6.04 | 6.26E-03 | 6.87 | 3.02E-05 |
| Glyceraldehyde | C02154 | HMDB0001051 | 0.29 | 5.86E-04 | 0.32 | 1.65E-03 |
| Sphingosine | C00319 | HMDB0000252 | 2.03 | 3.30E-02 | 3.34 | 2.18E-05 |
| Glycerophosphocholine | C00670 | HMDB0000086 | 56.88 | 1.93E-03 | 51.03 | 3.44E-03 |
| Glycine | C00037 | HMDB0000123 | 0.55 | 2.22E-02 | 0.47 | 4.68E-03 |
| Guanidine | C17349 | HMDB0001842 | 0.47 | 5.25E-03 | 0.01 | 3.79E-04 |
| Hexadecanedioic acid | C19615 | HMDB0000672 | 0.27 | 1.34E-02 | 0.23 | 3.44E-03 |
| Histamine | C00388 | HMDB0000870 | 0.58 | 2.49E-02 | 0.35 | 7.49E-03 |
| Hypotaurine | C00519 | HMDB0000965 | 0.07 | 4.19E-04 | 0.08 | 5.33E-04 |
| Inosinic acid | C00130 | HMDB0000175 | 92.72 | 1.73E-05 | 150.57 | 1.43E-03 |
| L-Carnitine | C00318 | HMDB0000062 | 2.24 | 2.39E-03 | 2.70 | 3.97E-03 |
| L-Cystine | C00491 | HMDB0000192 | 0.49 | 2.70E-02 | 0.13 | 1.17E-03 |
| L-Phenylalanine | C00079 | HMDB0000159 | 0.51 | 2.80E-02 | 0.31 | 4.70E-03 |
| N-Acetylneuraminic acid | C19910 | HMDB0000230 | 2.68 | 2.13E-02 | 2.90 | 3.38E-02 |
| Oxidized glutathione | C00127 | HMDB0003337 | 13.97 | 4.36E-03 | 16.05 | 2.77E-03 |
| L-Palmitoylcarnitine | C02990 | HMDB0000222 | 3.59 | 1.88E-02 | 5.09 | 2.77E-04 |
| Pantothenol | C05944 | HMDB0004231 | 0.44 | 3.12E-02 | 0.18 | 8.44E-05 |
| Pyroglutamic acid | C01879 | HMDB0000267 | 0.29 | 1.85E-03 | 0.16 | 5.30E-04 |
| Quinic acid | C06746 | HMDB0003072 | 0.09 | 4.05E-02 | 0.25 | 4.76E-02 |
| Retinal | C00376 | HMDB0001358 | 0.13 | 4.98E-02 | 0.43 | 3.81E-01 |
| Rhamnose | C00507 | HMDB0000849 | 0.22 | 2.90E-03 | 0.17 | 2.40E-04 |
| Deoxycholic acid glycine conjugate | C05464 | HMDB0000631 | 0.19 | 6.50E-03 | 0.15 | 4.52E-04 |
| Sorbitol | C00794 | HMDB0000247 | 0.29 | 9.19E-03 | 0.13 | 1.44E-03 |
| Tetradecanedioic acid | C11002 | HMDB0000872 | 0.44 | 4.46E-02 | 0.36 | 4.47E-02 |
| Thiamine | C00378 | HMDB0000235 | 0.34 | 6.95E-05 | 0.18 | 2.47E-05 |
In the fold change calculation, the metabolite in pre-gemcitabine normal tissues served as the denominator. Bold print indicates statistical significance. BCa, bladder cancer; KEGG, Kyoto Encyclopedia of Genes and Genomes; HMDB, human metabolome database.
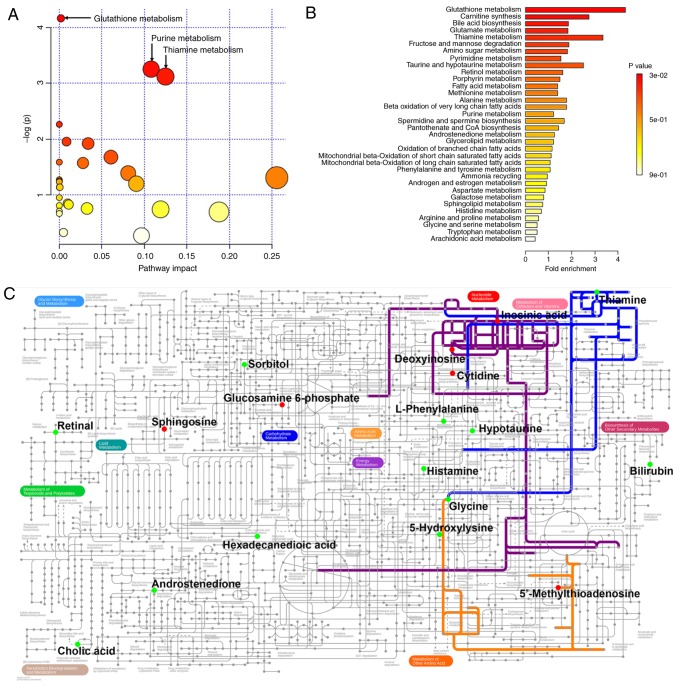
The 34 BCa-associated metabolites were then submitted to MetaboAnalyst for analysis. Table II lists all metabolites found to be altered in BCa. Table III and Fig. 2A show the metabolic pathways connected with these 34 metabolites, among which three pathways, namely glutathione, purine and thiamine metabolism, were significantly associated with BCa. Furthermore, in order to expand our understanding of metabolic pathways related to BCa, the module of enrichment analysis of MetaboAnalyst was used, which verified that glutathione metabolism was significantly associated with BCa (Table IV and Fig. 2B). The metabolic network of the differential metabolites and altered metabolic pathways in the KEGG general metabolic pathway map is shown in Fig. 2C.
Table III.
Pathway analysis of metabolite changes in BCaa.
| KEGG pathway | Total | Hits | P-value |
|---|---|---|---|
| Glutathione metabolism | 38 | 3 | 1.55E-02 |
| Purine metabolism | 92 | 4 | 3.86E-02 |
| Thiamine metabolism | 24 | 2 | 4.40E-02 |
| Nitrogen metabolism | 39 | 2 | 1.04E-01 |
| Primary bile acid biosynthesis | 47 | 2 | 1.41E-01 |
| Fructose and mannose metabolism | 48 | 2 | 1.46E-01 |
| Cysteine and methionine metabolism | 56 | 2 | 1.86E-01 |
| Cyanoamino acid metabolism | 16 | 1 | 2.04E-01 |
| Pyrimidine metabolism | 60 | 2 | 2.07E-01 |
| Taurine and hypotaurine metabolism | 20 | 1 | 2.48E-01 |
| Retinol metabolism | 22 | 1 | 2.70E-01 |
| Ether lipid metabolism | 23 | 1 | 2.80E-01 |
| Aminoacyl-tRNA biosynthesis | 75 | 2 | 2.86E-01 |
| Alanine, aspartate and glutamate metabolism | 24 | 1 | 2.90E-01 |
| Sphingolipid metabolism | 25 | 1 | 3.01E-01 |
| Pantothenate and CoA biosynthesis | 27 | 1 | 3.20E-01 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 27 | 1 | 3.20E-01 |
| Methane metabolism | 34 | 1 | 3.86E-01 |
| Glycerophospholipid metabolism | 39 | 1 | 4.28E-01 |
| Porphyrin and chlorophyll metabolism | 104 | 2 | 4.36E-01 |
| Galactose metabolism | 41 | 1 | 4.45E-01 |
| Histidine metabolism | 44 | 1 | 4.68E-01 |
| Phenylalanine metabolism | 45 | 1 | 4.76E-01 |
| Lysine degradation | 47 | 1 | 4.91E-01 |
| Glycine, serine and threonine metabolism | 48 | 1 | 4.98E-01 |
| Fatty acid metabolism | 50 | 1 | 5.13E-01 |
| Amino sugar and nucleotide sugar metabolism | 88 | 1 | 7.21E-01 |
| Steroid hormone biosynthesis | 99 | 1 | 7.63E-01 |
The analysis was conducted by the module of pathway analysis of MetaboAnalyst 4.0. BCa, bladder cancer; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 2.
Results of the metabolic connection analysis of the changed metabolomic data in BCa. (A) Pathway analysis based on the KEGG database. (B) Enrichment analysis based on SMPDB. (C) Metabolic network of the differential metabolites and altered metabolic pathways in KEGG general metabolic pathway map. Red dots represent the increased metabolites in BCa; green dots represent the specifically decreased metabolites in BCa; orange line, glutathione metabolism; purple line, purine metabolism; blue line, thiamine metabolism. The metabolites and pathways not indicated in the general pathway map are not shown. The original general metabolic pathway map is available at https://pathways.embl.de/ipath3.cgi. KEGG, Kyoto Encyclopedia of Genes and Genomes; SMPDB, Small Molecule Pathway Database; BCa, bladder cancer.
Table IV.
Enrichment analysis of metabolite changes in BCaa.
| Pathway from SMPDB | Total | Hits | P-value |
|---|---|---|---|
| Glutathione metabolism | 21 | 3 | 2.96E-02 |
| Carnitine synthesis | 22 | 2 | 1.64E-01 |
| Bile acid biosynthesis | 65 | 4 | 1.64E-01 |
| Glutamate metabolism | 49 | 3 | 2.19E-01 |
| Thiamine metabolism | 9 | 1 | 2.63E-01 |
| Fructose and mannose degradation | 32 | 2 | 2.88E-01 |
| Amino Sugar metabolism | 33 | 2 | 3.00E-01 |
| Pyrimidine metabolism | 59 | 3 | 3.11E-01 |
| Taurine and hypotaurine metabolism | 12 | 1 | 3.35E-01 |
| Retinol metabolism | 37 | 2 | 3.50E-01 |
| Porphyrin metabolism | 40 | 2 | 3.87E-01 |
| Fatty acid metabolism | 43 | 2 | 4.23E-01 |
| Methionine metabolism | 43 | 2 | 4.23E-01 |
| Alanine metabolism | 17 | 1 | 4.39E-01 |
| Beta oxidation of very long-chain fatty acids | 17 | 1 | 4.39E-01 |
| Purine metabolism | 74 | 3 | 4.51E-01 |
| Spermidine and spermine biosynthesis | 18 | 1 | 4.58E-01 |
| Pantothenate and CoA biosynthesis | 21 | 1 | 5.11E-01 |
| Androstenedione metabolism | 24 | 1 | 5.59E-01 |
| Glycerolipid metabolism | 25 | 1 | 5.74E-01 |
| Oxidation of branched chain fatty acids | 26 | 1 | 5.89E-01 |
| Mitochondrial beta-oxidation of short-chain saturated fatty acids | 27 | 1 | 6.03E-01 |
| Mitochondrial beta-oxidation of long-chain saturated fatty acids | 28 | 1 | 6.16E-01 |
| Phenylalanine and tyrosine metabolism | 28 | 1 | 6.16E-01 |
| Ammonia recycling | 32 | 1 | 6.66E-01 |
| Androgen and estrogen metabolism | 33 | 1 | 6.78E-01 |
| Aspartate metabolism | 35 | 1 | 6.99E-01 |
| Galactose metabolism | 38 | 1 | 7.29E-01 |
| Sphingolipid metabolism | 40 | 1 | 7.48E-01 |
| Histidine metabolism | 43 | 1 | 7.73E-01 |
| Arginine and proline metabolism | 53 | 1 | 8.41E-01 |
| Glycine and serine metabolism | 59 | 1 | 8.72E-01 |
| Tryptophan metabolism | 60 | 1 | 8.76E-01 |
| Arachidonic acid metabolism | 69 | 1 | 9.10E-01 |
The analysis was conducted by the module of enrichment analysis of MetaboAnalyst 4.0. BCa, bladder cancer. SMPDB, Small Molecule Pathway Database.
Candidate targets of submucosal injection of gemcitabine
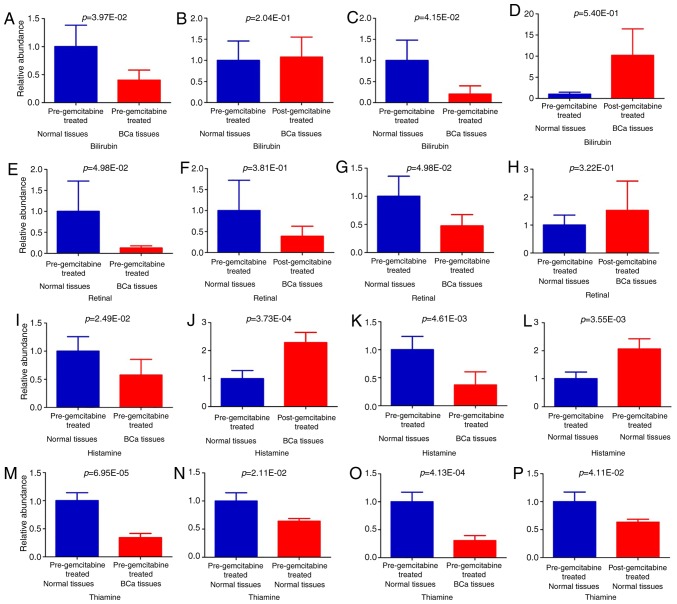
Based on targeted metabolomic analysis, we next analyzed these 34 differential metabolite changes in post-gemcitabine BCa tissues, and compared their levels with those in pre-gemcitabine normal tissues. A total of 32 metabolites maintained significant changes with identical trends in the comparison between pre-gemcitabine normal vs. pre-gemcitabine BCa tissues, indicating that the findings for differential metabolites in BCa are reliable. Importantly, the significant decrease in the levels of two metabolites associated with BCa recovered to insignificant levels following submucosal injection of gemcitabine (Table II and Fig. 3A, B and E-F). These were bilirubin and retinal, which may be the candidate targets of gemcitabine for the prevention of recurrence of urothelial BCa. We further analyzed the changes in the two metabolites in association with cancer stage. In Ta/T1 stage disease, when comparing pre-gemcitabine normal vs. pre-gemcitabine BCa tissues, bilirubin was decreased significantly in the BCa tissues (P=4.15E-2) (Table SI and Fig. 3C), while this decrease recovered to an insignificant level following submucosal injection of gemcitabine (P=5.40E-1) (Table SI and Fig. 3D); however, at this stage, the decrease of retinal in the tumor was still not statistically significant (P=4.76E-1) (Table SI). In T2 stage disease, when comparing pre-gemcitabine normal vs. pre-gemcitabine BCa tissues, retinal was decreased significantly in the BCa tissues (P=4.98E-2) (Table SI and Fig. 3G), while its change became statistically insignificant following submucosal injection of gemcitabine (P=3.22E-1) (Table SI and Fig. 3H); however, at this stage, the decrease of retinal in tumor was not statistically significant (P=1.68E-1) (Table SI). These results indicate that bilirubin and retinal changes were correlated with cancer stage, and gemcitabine may exert its effect through metabolic pathways associated with cancer stage.
Figure 3.
Relative levels of potential candidate targets of submucosal injection of gemcitabine (A, B, E, F, I, J, M and N). All samples; (C and D) Ta/T1 stage; (G, H, K, L, O and P) T2 stage. Values are presented as mean ± standard error of the mean. The P-values of paired t-test are indicated.
Effects of submucosal injection of gemcitabine on normal tissues
To identify the effects of submucosal injection of gemcitabine on normal bladder tissues, the metabolomes of normal tissues were compared pre- and post-gemcitabine treatment. A total of 10 metabolites were found to be significantly altered in the normal tissues following submucosal injection of gemcitabine, whereas only 2 metabolites were associated with BCa (Table V). Histamine was significantly decreased in BCa, and its level was significantly increased in normal tissues after submucosal injection of gemcitabine (Fig. 3I and J). However, thiamine was significantly decreased in BCa, and its level was significantly decreased in normal tissues after submucosal injection of gemcitabine (Fig. 3M and N). We further analyzed the changes in the two metabolite in relation to cancer stage. In Ta/T1 stage disease, when comparing pre-gemcitabine normal vs. pre-gemcitabine BCa tissues, the changes in histamine (P=2.60E-1) and thiamine (P=8.39E-2) in the tumor were insignificant (Table SI). However, in T2 stage disease, when comparing pre-gemcitabine normal vs. pre-gemcitabine BCa tissues, the decrease in histamine (P=4.61E-3) (Table SI and Fig. 3K) and thiamine (P=4.13E-4) (Table SI and Fig. 3O) in the BCa tissues was significant. In T2 stage disease, the histamine level was significantly increased in normal tissues after submucosal injection of gemcitabine (P=3.55E-3) (Table SI and Fig. 3L), while thiamine was significantly decreased (P=4.11E-2) (Table SI and Fig. 3P). These results indicate that these metabolite changes were correlated with cancer stage, and gemcitabine may exert its effects mainly through these metabolic pathways in T2 stage disease.
Table V.
Changes in normal tissues after gemcitabine treatment.
| Metabolites | Pre-gemcitabine BCa vs. normal tissuesa | Post-gemcitabine vs. pre-gemcitabine normal tissuesa | ||
|---|---|---|---|---|
| Fold-changea | P-value | Fold-changea | P-value | |
| 3-Methyladenine | 1.54 | 3.78E-01 | 0.17 | 2.33E-02 |
| Ascorbic acid | 22.26 | 1.85E-01 | 0.28 | 5.46E-03 |
| Creatinine | 0.48 | 3.44E-01 | 0.13 | 1.16E-02 |
| D-Glyceraldehyde 3-phosphate | 0.72 | 3.33E-01 | 0.15 | 2.16E-02 |
| Histamine | 0.58 | 2.49E-02 | 2.26 | 3.73E-04 |
| N-Acetylglutamic acid | 0.52 | 8.58E-02 | 0.57 | 4.95E-02 |
| N-Acetylglutamine | 0.68 | 5.45E-01 | 0.11 | 4.25E-02 |
| Thiamine | 0.34 | 6.95E-05 | 0.65 | 2.11E-02 |
| Tryptamine | 0.10 | 7.66E-02 | 0.11 | 3.23E-02 |
| Uridine | 1.41 | 6.74E-02 | 1.51 | 3.65E-02 |
In the fold change calculation, the metabolite in pre-gemcitabine normal tissues served as the denominator. Bold print indicates statistically significant differences. BCa, bladder cancer.
Discussion
In the present study, UPLC-Q-Exactive-based metabolomic analysis was utilized to profile metabolites in BCa tissues. The major findings may be summarized as follows: i) A total of 34 key metabolites associated with BCa were identified (Table II); ii) three metabolic pathways, namely glutathione, purine and thiamine metabolism, were altered in BCa, and glutathione metabolism was the consistently altered pathway in enrichment and pathway analyses (Tables III and IV, and Fig. 2A and B); iii) among the 34 cancer-associated metabolites, the levels of bilirubin and retinal recovered after gemcitabine injection, suggesting that these two are likely the targets of gemcitabine treatment (Table II, Fig. 3A-D and E-H); iv) the effects of gemcitabine on normal bladder tissues were also investigated, and it was deduced that histamine may have the ability to protect against disease recurrence, whereas thiamine may be involved in the side effects of treatment (Table V, Fig. 3I-L and M-P), which requires further confirmation in future studies.
The identification of the three metabolic pathways significantly altered in BCa may be pathophysiologically important. Glycine is an important amino acid that participates in all three metabolic pathways. A decrease in glycine was reported as a biomarker in BCa (14). As a precursor of purine synthesis, reduced glycine in BCa indicates that a critical metabolic process associated with cell proliferation is altered in BCa (14). Notably, glutathione metabolism is the consistently altered pathway in enrichment and pathway analysis. Glutathione is the most abundant low-molecular-weight peptide present in eukaryotic cells (28). Glutathione is a primary cellular antioxidant that effectively scavenges free radicals and other reactive oxygen species (29) and, therefore, plays an important role in protecting cells from oxidative injury (30). Glutathione is also involved in cellular detoxification, and is required in several aspects of the immune response (31). Ke et al reported altered oxidized glutathione in BCa; they discovered four single-nucleotide polymorphisms in the glutathione synthetase gene, and these changes were associated with BCa recurrence after TUR and Bacillus Calmette Guerin treatment (31). Our findings, together with others, suggest that oxidative stress in BCa cells is at least partially due to the disrupted glutathione metabolism.
By using LC-Q-Exactive MS, this study is, to the best of our knowledge, the first to investigate the metabolite changes in BCa treated with gemcitabine followed by TURBT. Among the 34 cancer-associated metabolites, the levels of bilirubin and retinal recovered after gemcitabine treatment, indicating that they may be the potential targets of gemcitabine for reducing BCa recurrence. Bilirubin, a degradation product of free heme groups, protected LLC-PK1 cells against cisplatin-induced death (32). A large population-based study demonstrated that patients with primary biliary cirrhosis (PBC) have a nine-fold increased risk of developing urinary BCa, and BCa and PBC share a number of etiological factors (33). Uridine 5'-diphospho-glucuronosyltransferases (UGTs) are enzymes that participate in several biological processes involving bilirubin conjugation. UGTs catabolize carcinogens and, therefore, protect bladder cells from the harmful effects of toxic chemicals accumulated in the bladder. Targeting UGT1A may serve as a novel therapeutic intervention against uroepithelial carcinomas (34). Retinal (retinaldehyde; Rald) is derived from retinol (vitamin A) oxidized by alcohol dehydrogenases (35). Retinal plays an essential role in molecular signaling in vision, and serves mainly as a retinoic acid (an active form of vitamin A) precursor outside the eye (36). The serum levels of vitamin A were found to be decreased in patients with BCa (37). A high dietary intake of vitamin A reduces the incidence of BCa (38). Ziouzenkova et al reported retinal as a distinct biological regulator involved in suppressing adipo-genesis, diet-induced obesity and insulin resistance (39). The potential effect of bilirubin and retinal on the clinical outcome of patients with BCa identified in the present study is worthy of further investigation in the future.
The effects of gemcitabine on the metabolism of adjacent normal tissues were also examined. Most gemcitabine-induced metabolites do not overlap with those identified in BCa, indicating that gemcitabine did not exert notable adverse effects on normal bladder tissues. It was observed that histamine change may be associated with the prevention of relapse. Histamine is derived from the decarboxylation of histidine by histidine decarboxylase in mammals (8). Histamine is primarily released in inflammatory processes by mast cells (8), which are closely associated with BCa (40). Histamine H1 receptor (HRH1) expression was identified in BCa and found to be associated with the prognosis (41). It was observed that thiamine change may be involved in treatment-related side effects. Thiamine, or vitamin B-1, is a water-soluble vitamin (42). An early report by Pamukcu et al demonstrated that the incidence of urinary bladder carcinomas in rats fed bracken fern and additionally s.c. injected once weekly with 2 mg of thiamine hydrochloride was significantly higher compared with that in rats fed bracken fern but receiving no thiamine supplements, as thiamine may interfere with the absorption, distribution, metabolism, or excretion of the bracken fern (43). However, a case-control study from New Hampshire investigated the effect of minerals and vitamins on the risk of BCa, and found that a higher total intake of thiamine was inversely correlated with BCa risk in older participants (44).
There is currently a lack of effective biomarkers for BCa diagnosis and prognosis. Metabolomic profiles from tissue have the potential to be used, along with other current diagnostics, to help guide the clinical management of patients with BCa. The changed metabolites identified in the present study, such as glycine, may be used as potential biomarkers for BCa. In addition, those metabolites discovered in BC gemcitabine treatment may reveal new metabolic pathways that mediate the anti-recurrence effect of gemcitabine, which currently remain elusive.
In summary, the present study employed UPLC-HRMS-based metabolomic analysis to investigate metabolite changes in bladder tissues from BCa and BCa treated with gemcitabine. The findings may provide new insights into metabolic changes in BCa and the biomolecular basis of submucosal injection of gemcitabine for BCa. In addition, the study demonstrated that the UPLC-HRMS-based metabolomic analysis provides comprehensive metabolite profiling data that may pave the way to a novel approach to BCa research.
Supplementary Data
Acknowledgments
Not applicable.
Abbreviations
- TURBT
transurethral resection of bladder tumor
- PCA
principal component analysis
- ACN
acetonitrile
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- HMDB
human metabolome database
- SMPDB
Small Molecule Pathway Database
- HPLC/MS
high-performance liquid chromatography/ mass spectrometry
- UPLC-MS
ultra-performance LC-MS
- UPLC-TOF-MS
UPLC time-of-flight MS
- UPLC-HRMS
UPLC high-resolution MS
- BCa
bladder cancer
Funding
The present study was supported by grants from the National Natural Science Foundation (81872650 and 81573182); the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (18KJA320003 and 18KJB320001); the Key Research & Development Plan of Jiangsu Province (BE2017628); the Southeast University & Nanjing Medical University Collaborative Research Project (2242018K3DN25); the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD); the 333 Project of Jiangsu Province (BRA2017241); the Natural Science Foundation of Huai'an (HAB201801); the Huai'an Promotion Project for Science and Technology International Cooperation (HAC201708); and the Innovation Fund Project of the State Key Laboratory of Reproductive Medicine (SKLRM-GC201901).
Availability of data and materials
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
CY, XS, KW, YG, RS and MC designed and performed the experiments. TL and MC analyzed and interpreted the raw data. SZ, XJ, BZ and HW collected the bladder tissue samples. ZG and SS performed the histopathological examination of BCa tissues. JL and JT preserved the samples and collected the basic information of the participants. CY, TL and MC wrote the manuscript. XW, HJ, XN, XW, MC and GF critically revised the manuscript for important intellectual content. MC and GF supervised the project. All authors have read and approved the final version of the manuscript for publication.
Ethics approval and consent to participate
The Ethics Committee of The Affiliated Huai'an No. 1 People's Hospital of Nanjing Medical University reviewed and approved the study protocol (serial no. YL-P-2013-21-01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Authors' information
Chao Yang: 15949177271@163.com; Xian Sun: 1019949991@qq.com; Hengbing Wang: wanghengbing2004@163.com; Ting Lu: 552095014@qq.com; Keqing Wu: wkwly1993@qq.com; Yusheng Guan: guanys@njmu.edu.cn; Jing Tang: tjing19681222@sina.com; Jian Liang: lj3936@126.com; Rongli Sun: 101012172@seu.edu.cn; Zhongying Guo: guozhongying407@163.com; Sinian Zheng: zhengsinian@163.com; Xiaoli Wu: wuxiaoli2233@126.com; Hesong Jiang: njjhs2007@163.com; Xi Jiang: piscesjxi@163.com; Bing Zhong: 15152569186@163.com; Xiaobing Niu: bingke2008@sina.com; Suan Sun: hayyssa@163.com; Xinru Wang: xrwang@njmu.edu.cn
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Messing EM, Madeb R, Young T, Gilchrist KW, Bram L, Greenberg EB, Wegenke JD, Stephenson L, Gee J, Feng C. Long-term outcome of hematuria home screening for bladder cancer in men. Cancer. 2006;107:2173–2179. doi: 10.1002/cncr.22224. [DOI] [PubMed] [Google Scholar]
- 3.Messing EM, Tangen CM, Lerner SP, Sahasrabudhe DM, Koppie TM, Wood DP, Jr, Mack PC, Svatek RS, Evans CP, Hafez KS, et al. Effect of intravesical instillation of gemcitabine vs saline immediately following resection of suspected low-grade non-muscle-invasive bladder cancer on tumor recurrence: SWOG S0337 randomized clinical trial. JAMA. 2018;319:1880–1888. doi: 10.1001/jama.2018.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Wang B, Tian HZ, Gao JZ. Submucosal injection of anti-tumor drug on the prevention of Post-TUR-Bt recurrence. Zhonghua Wai Ke Za Zhi. 2004;42:580–582. In Chinese. [PubMed] [Google Scholar]
- 5.Caffo O, Thompson C, De Santis M, Kragelj B, Hamstra DA, Azria D, Fellin G, Pappagallo GL, Galligioni E, Choudhury A. Concurrent gemcitabine and radiotherapy for the treatment of muscle-invasive bladder cancer: A pooled individual data analysis of eight phase I-II trials. Radiother Oncol. 2016;121:193–198. doi: 10.1016/j.radonc.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Sternberg CN, Bellmunt J, Sonpavde G, Siefker-Radtke AO, Stadler WM, Bajorin DF, Dreicer R, George DJ, Milowsky MI, Theodorescu D, et al. ICUD-EAU international consultation on bladder cancer 2012: Chemotherapy for urothelial carcinoma-neoadjuvant and adjuvant settings. Eur Urol. 2013;63:58–66. doi: 10.1016/j.eururo.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Van QN, Veenstra TD. How close is the bench to the bedside? Metabolic profiling in cancer research. Genome Med. 2009;1:5. doi: 10.1186/gm5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bansal N, Gupta A, Mitash N, Shakya PS, Mandhani A, Mahdi AA, Sankhwar SN, Mandal SK. Low- and high-grade bladder cancer determination via human serum-based metabolo-mics approach. J Proteome Res. 2013;12:5839–5850. doi: 10.1021/pr400859w. [DOI] [PubMed] [Google Scholar]
- 9.Jin X, Yun SJ, Jeong P, Kim IY, Kim WJ, Park S. Diagnosis of bladder cancer and prediction of survival by urinary metabo-lomics. Oncotarget. 2014;5:1635–1645. doi: 10.18632/oncotarget.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Issaq HJ, Nativ O, Waybright T, Luke B, Veenstra TD, Issaq EJ, Kravstov A, Mullerad M. Detection of bladder cancer in human urine by metabolomic profiling using high performance liquid chromatography/mass spectrometry. J Urol. 2008;179:2422–2426. doi: 10.1016/j.juro.2008.01.084. [DOI] [PubMed] [Google Scholar]
- 11.Pasikanti KK, Esuvaranathan K, Hong Y, Ho PC, Mahendran R, Raman Nee Mani L, Chiong E, Chan EC. Urinary metabotyping of bladder cancer using two-dimensional gas chromatography time-of-flight mass spectrometry. J Proteome Res. 2013;12:3865–3873. doi: 10.1021/pr4000448. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Cheng X, Liu X, He L, Zhang W, Wang Y, Sun W, Ji Z. Investigation of the urinary metabolic variations and the application in bladder cancer biomarker discovery. Int J Cancer. 2018;143:408–418. doi: 10.1002/ijc.31323. [DOI] [PubMed] [Google Scholar]
- 13.Alberice JV, Amaral AF, Armitage EG, Lorente JA, Algaba F, Carrilho E, Márquez M, García A, Malats N, Barbas C. Searching for urine biomarkers of bladder cancer recurrence using a liquid chromatography-mass spectrometry and capillary electrophoresis-mass spectrometry metabolomics approach. J Chromatogr A. 2013;1318:163–170. doi: 10.1016/j.chroma.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, Song R, Ma C, Zhou L, Liu X, Yin P, Zhang Z, Sun Y, Xu C, Lu X, Xu G. Discovery and validation of potential urinary biomarkers for bladder cancer diagnosis using a pseudotargeted GC-MS metabolomics method. Oncotarget. 2017;8:20719–20728. doi: 10.18632/oncotarget.14988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao CH, Chen CL, Lin JY, Chen CJ, Fu SH, Chen YT, Chang YS, Yu JS, Tsui KH, Juo CG, Wu KP. Metabolite marker discovery for the detection of bladder cancer by comparative metabolomics. Oncotarget. 2017;8:38802–38810. doi: 10.18632/oncotarget.16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Rundstedt FC, Rajapakshe K, Ma J, Arnold JM, Gohlke J, Putluri V, Krishnapuram R, Piyarathna DB, Lotan Y, Gödde D, et al. Integrative pathway analysis of metabolic signature in bladder cancer: A linkage to the cancer genome atlas project and prediction of survival. J Urol. 2016;195:1911–1919. doi: 10.1016/j.juro.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Putluri N, Shojaie A, Vasu VT, Vareed SK, Nalluri S, Putluri V, Thangjam GS, Panzitt K, Tallman CT, Butler C, et al. Metabolomic profiling reveals potential markers and biopro-cesses altered in bladder cancer progression. Cancer Res. 2011;71:7376–7386. doi: 10.1158/0008-5472.CAN-11-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Y, Yang X, Deng X, Zhang X, Li P, Tao J, Qin C, Wei J, Lu Q. Metabolomics in bladder cancer: A systematic review. Int J Clin Exp Med. 2015;8:11052–11063. [PMC free article] [PubMed] [Google Scholar]
- 19.Eidelman E, Tripathi H, Fu DX, Siddiqui MM. Linking cellular metabolism and metabolomics to risk-stratification of prostate cancer clinical aggressiveness and potential therapeutic pathways. Transl Androl Urol. 2018;7(Suppl 4):S490–S497. doi: 10.21037/tau.2018.04.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lima AR, Bastos Mde L, Carvalho M, Guedes de Pinho P. Biomarker discovery in human prostate cancer: An update in metabolomics studies. Transl Oncol. 2016;9:357–370. doi: 10.1016/j.tranon.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein JI, Amin MB, Reuter VR, Mostofi FK. The world health organization/international society of urological pathology consensus classification of urothelial (transitional cell) neoplasms of the urinary bladder. Bladder consensus conference committee Am J Surg Pathol. 1998;22:1435–1448. doi: 10.1097/00000478-199812000-00001. [DOI] [PubMed] [Google Scholar]
- 22.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 23.Hu W, Dong T, Wang L, Guan Q, Song L, Chen D, Zhou Z, Chen M, Xia Y, Wang X. Obesity aggravates toxic effect of BPA on spermatogenesis. Environ Int. 2017;105:56–65. doi: 10.1016/j.envint.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Christen S, Lorendeau D, Schmieder R, Broekaert D, Metzger K, Veys K, Elia I, Buescher JM, Orth MF, Davidson SM, et al. Breast cancer-derived lung metastases show increased pyruvate carboxylase-dependent anaplerosis. Cell Rep. 2016;17:837–848. doi: 10.1016/j.celrep.2016.09.042. [DOI] [PubMed] [Google Scholar]
- 25.Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O'Brien JP, Pierce KA, et al. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, et al. SMPDB: The small molecule pathway database. Nucleic Acids Res. 2010;38:D480–D487. doi: 10.1093/nar/gkp1002. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He YT, Li DJ, Liang D, Zheng RS, Zhang SW, Zeng HM, Chen WQ, He J. Incidence and mortality of bladder cancer in China, 2014. Zhonghua Zhong Liu Za Zhi. 2018;40:647–652. doi: 10.3760/cma.j.issn.0253-3766.2018.09.002. In Chinese. [DOI] [PubMed] [Google Scholar]
- 28.Estrela JM, Ortega A, Obrador E. Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci. 2006;43:143–181. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- 29.Balendiran GK, Dabur R, Fraser D. The role of glutathione in cancer. Cell Biochem Funct. 2004;22:343–352. doi: 10.1002/cbf.1149. [DOI] [PubMed] [Google Scholar]
- 30.Amelio I, Cutruzzolá F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191–198. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ke HL, Lin J, Ye Y, Wu WJ, Lin HH, Wei H, Huang M, Chang DW, Dinney CP, Wu X. Genetic variations in glutathione pathway genes predict cancer recurrence in patients treated with transurethral resection and bacillus calmetteguerin instillation for non-muscle invasive bladder cancer. Ann Surg Oncol. 2015;22:4104–4110. doi: 10.1245/s10434-015-4431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patricia Moreno-Londoño A, Bello-Alvarez C, Pedraza- Chaverri J. Isoliquiritigenin pretreatment attenuates cisplatin induced proximal tubular cells (LLC-PK1) death and enhances the toxicity induced by this drug in bladder cancer T24 cell line. Food Chem Toxicol. 2017;109:143–154. doi: 10.1016/j.fct.2017.08.047. [DOI] [PubMed] [Google Scholar]
- 33.Boonstra K, Bokelaar R, Stadhouders PH, Tuynman HA, Poen AC, van Nieuwkerk KM, Witteman EM, Hamann D, Witteman BJ, Beuers U, Ponsioen CY. Increased cancer risk in a large population-based cohort of patients with primary biliary cirrhosis: Follow-up for up to 36 years. Hepatol Int. 2014;8:266–274. doi: 10.1007/s12072-014-9530-z. [DOI] [PubMed] [Google Scholar]
- 34.Sundararaghavan VL, Sindhwani P, Hinds TD., Jr Glucuronidation and UGT isozymes in bladder: New targets for the treatment of uroepithelial carcinomas? Oncotarget. 2017;8:3640–3648. doi: 10.18632/oncotarget.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duester G, Mic FA, Molotkov A. Cytosolic retinoid dehydro-genases govern ubiquitous metabolism of retinol to retinaldehyde followed by tissue-specific metabolism to retinoic acid. Chem Biol Interact. 2003;143-144:201–210. doi: 10.1016/S0009-2797(02)00204-1. [DOI] [PubMed] [Google Scholar]
- 36.Napoli JL. Retinoic acid: Its biosynthesis and metabolism. Prog Nucleic Acid Res Mol Biol. 1999;63:139–188. doi: 10.1016/S0079-6603(08)60722-9. [DOI] [PubMed] [Google Scholar]
- 37.Mahmoud LA, Robinson WA. Vitamin A levels in human bladder cancer. Int J Cancer. 1982;30:143–145. doi: 10.1002/ijc.2910300203. [DOI] [PubMed] [Google Scholar]
- 38.Mettlin C, Graham S. Dietary risk factors in human bladder cancer. Am J Epidemiol. 1979;110:255–263. doi: 10.1093/oxfordjournals.aje.a112810. [DOI] [PubMed] [Google Scholar]
- 39.Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, Hamilton JA, Tang G, Dolnikowski GG, Vogel S, et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med. 2007;13:695–702. doi: 10.1038/nm1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serel TA, Soyupek S, Çandir Ö. Association between mast cells and bladder carcinoma. Urol Int. 2004;72:299–302. doi: 10.1159/000077681. [DOI] [PubMed] [Google Scholar]
- 41.Wang M, Wei X, Shi L, Chen B, Zhao G, Yang H. Integrative genomic analyses of the histamine H1 receptor and its role in cancer prediction. Int J Mol Med. 2014;33:1019–1026. doi: 10.3892/ijmm.2014.1649. [DOI] [PubMed] [Google Scholar]
- 42.Kerns JC, Gutierrez JL. Thiamin Adv Nutr. 2017;8:395–397. doi: 10.3945/an.116.013979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pamukcu AM, Yalciner S, Price JM, Bryan GT. Effects of the coadministration of thiamine on the incidence of urinary bladder carcinomas in rats fed bracken fern. Cancer Res. 1970;30:2671–2674. [PubMed] [Google Scholar]
- 44.Brinkman MT, Karagas MR, Zens MS, Schned A, Reulen RC, Zeegers MP. Minerals and vitamins and the risk of bladder cancer: Results from the new hampshire study. Cancer Causes Control. 2010;21:609–619. doi: 10.1007/s10552-009-9490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.