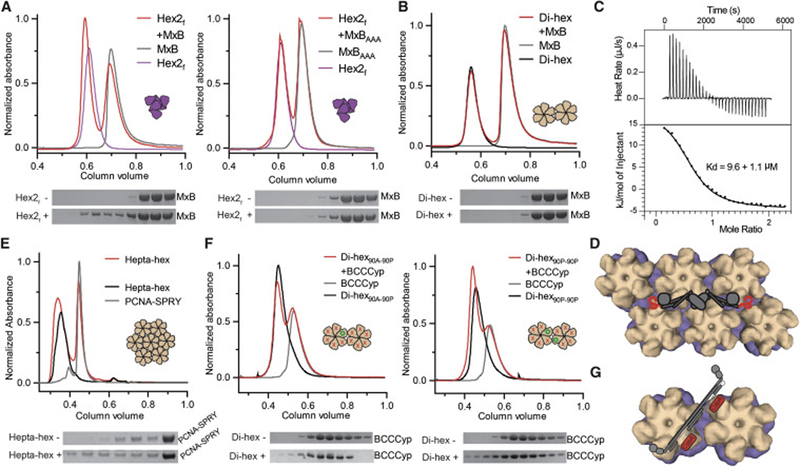
Figure 6. MxB and TRIM5 proteins recognize inter-hexamer interfaces.
(A) MxB1–83 (left), but not the 11AAA13 mutant (right), co-elutes with hexamer-2foldon (red) in SEC (top) and SDS-PAGE (bottom) analyses. (B) MxB1–83 does not co-elute with di-hexamers in SEC. (C) MxB1–83 binds to hexamer-2foldon with a 9.6 ± 1.1 μM Kd by ITC. (D) Model of full-length MxB (cartoon) wedging its unstructured N-termini into two disparate three-fold inter-hexamer interfaces (surface). (E) A PCNA trimer fused to the SPRY domain of rhesus TRIM5α co-elutes with hepta-hexamers (red) in SEC. (F) BCCCyp shows marginal binding to a di-hexamer containing only one wild-type P90 site (left), but significant binding to a di-hexamer with two wild-type P90 sites on adjacent hexamers (right). P90 is indicated with green circles and P90A as red crosses. (G) A model of flexible TRIMCyp (cartoon) binding between hexamers (surface), and within hexamers (Figure 2). See also Figure S6.