Abstract
The complex process of platelet formation originates with the hematopoietic stem cell, which differentiates through the myeloid lineage, matures, and releases proplatelets into the BM sinusoids. How formed platelets maintain a low basal activation state in the circulation remains unknown. We identify Lepr+ stromal cells lining the BM sinusoids as important contributors to sustaining low platelet activation. Ablation of murine Lepr+ cells led to a decreased number of platelets in the circulation with an increased activation state. We developed a potentially novel culture system for supporting platelet formation in vitro using a unique population of CD51+PDGFRα+ perivascular cells, derived from human umbilical cord tissue, which display numerous mesenchymal stem cell (MSC) properties. Megakaryocytes cocultured with MSCs had altered LAT and Rap1b gene expression, yielding platelets that are functional with low basal activation levels, a critical consideration for developing a transfusion product. Identification of a regulatory cell that maintains low baseline platelet activation during thrombopoiesis opens up new avenues for improving blood product production ex vivo.
Keywords: Hematology, Stem cells
Keywords: Human stem cells, Mouse stem cells, Platelets
Leptin receptor-positive stromal cells lining the bone marrow sinusoids are an important contributor to sustaining low platelet activation.
Introduction
Platelet formation in vivo occurs through an intricate series of events initiated from the hematopoietic stem cell (HSC), which undergoes differentiation through the myeloid lineage to the megakaryocyte progenitor stage and then matures to form proplatelets (1, 2). These extruded proplatelets are released into the BM sinusoids, after which the shear forces of the circulation break them off into individual platelets. Remarkably, the platelets released into the circulation have very low levels of activation. How formed platelets are able to maintain low activation levels in vivo remains a longstanding question.
Significant advances have been made to generate platelets in vitro as alternative sources of blood products. Nevertheless, while megakaryocytes are readily able to generate new platelets in vivo, this feature is limited in culture (3–5). Furthermore, existing methods have low efficiency and yield platelets with incomplete functionality at levels below those required for clinical use (4). These culture systems predominantly consist of cells seeded on a culture plate with static growth media containing a cocktail of cytokines or small molecules and, thereby, embody only a partial representation of the native microenvironment. Recently, the addition of shear stress or turbulent forces to culture systems has been demonstrated to increase platelet production (4, 6–8). While the VerMES bioreactor utilizing turbulent flow was able to generate significant quantities of platelets, it was not demonstrated whether the platelets generated with this device contained a microtubule coil, suggesting that they may be activated at baseline (8). Thus, there is a need to improve current in vitro platelet production approaches to generate platelets with a low resting state. Cell signaling in megakaryocyte progenitors through direct or indirect contact with other cells natively found in the in vivo microenvironment has not been sufficiently investigated and may be the key to generating platelets in vitro with low levels of baseline activation.
Much effort has been focused on characterizing the HSC niche, but considerably less is known about the key cells and microenvironment necessary for megakaryocyte differentiation and platelet formation. The BM sinusoids are lined with populations of stromal cells overlapping in expression of CXCL12, stem cell factor (SCF), leptin receptor (Lepr), Nestin, Osterix (OSX), and paired related homeobox-1 (Prx-1) (9–11). Perivascular cells marked by Nestin and overlapping with CD51+PDGFRα+ expression contain all of the mesenchymal stem cell (MSC) activity of the BM, form a unique niche with HSCs, and actively play a role in regulating HSC mobilization from the BM (12, 13). Murine megakaryocytes are thought to receive signals from endothelial or stromal cells lining the BM sinusoids, which promote megakaryocyte migration to the sinusoids (14). However, the population of cells responsible for providing these signaling cues has not been definitively identified. Furthermore, whether MSCs may play a role in regulating HSCs differentiated into megakaryocytes has not been well investigated. Previously, it was shown that human BM MSCs (bMSCs), defined by their adherence to tissue culture plates, can improve platelet formation in vitro, although the nature of the cells was not well defined (15). Human umbilical cord (UC) tissue, has also been shown to harbor MSCs, although these cells have not been thoroughly characterized (16–21).
In this study, we describe the potentially novel role of Lepr+ perisinusoidal stromal cells in assisting platelets to maintain low levels of basal activation in the circulation in vivo. Furthermore, we identify and rigorously characterize a population of CD51+PDGFRα+ perivascular stromal cells found around human umbilical arteries with similar platelet-promoting properties. Specifically, we demonstrate that these cells can not only enhance platelet formation in culture, but also importantly lead to the production of platelets with low basal activation, which are nevertheless functional following stimulation. The identification of MSCs for their ability to increase platelet numbers that are in a lower activation state expands the knowledge base of the megakaryocyte cell microenvironment, initiates drug development to target platelet-mediated inflammation or disorders of platelet activation, and potentially leads to the development of improved methods for ex vivo platelet production.
Results
Lepr+ perisinusoidal stromal cells assist in maintaining low basal platelet activation in vivo.
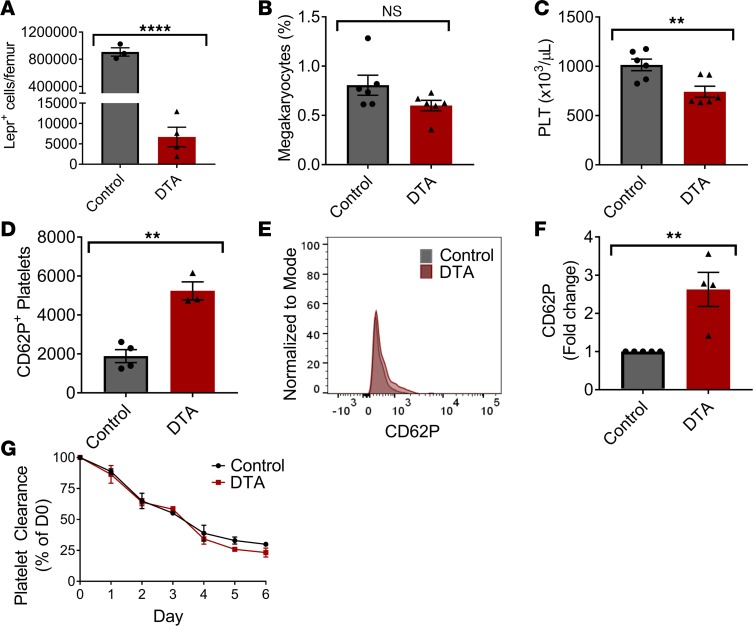
We investigated the role of stromal cells in reducing activation of platelets in vivo. Murine megakaryocytes are thought to receive signals from endothelial or stromal cells lining the BM sinusoids, which promote megakaryocyte migration to the sinusoids. However, the effect of the stromal cells on lowering platelet activation has not been investigated. We generated a triple transgenic mouse model containing Lepr, tdTomato, and diphtheria toxin fragment A (DTA) expression, which — upon recombination — lead to the depletion of tdTomato-labeled Lepr+ cells lining the BM sinusoids. Expression of tdTomato in Lepr+ cells was used to evaluate the level of DTA-mediated depletion. The DTA mice appeared normal in behavior, had normal skeletal formation, and did not have any differences in weights at 3 or 11 weeks of age (Supplemental Figure 1I; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.126982DS1) compared with the control mice. We confirmed the depletion of Lepr+ cells in the DTA group compared with the control group (Figure 1A). Interestingly, there was no significant change in the number of megakaryocytes found in the BM (Figure 1B). A complete blood count with differential revealed a significant decrease in the platelet number in the DTA group compared with the control (Figure 1C) but no other significant changes among other blood cell types (Supplemental Figure 1, A–H). We did not observe any changes in platelet α-granules at baseline, as assessed by intracellular vWF levels (Supplemental Figure 2A). Furthermore, platelets from the DTA group did not have any significant changes in their levels of phosphorylated serine or phosphorylated tyrosine at baseline (Supplemental Figure 2, B and C). However, we observed a significant increase in the basal levels of CD62P+-activated platelets in the BM compared with the control group (Figure 1, D–F), along with increased levels of integrin αIIβ3 (Supplemental Figure 2D). We did not observe any significant difference in the clearance of platelets in the DTA group compared with the control group (Figure 1G), affirming previous studies demonstrating that platelet activation does not affect the rate of platelet clearance (22). Platelets from the DTA mice were able to activate in response to 80 μg/ml collagen, 1 U/ml thrombin, and 20 μM ADP (Supplemental Figure 3, A–C) and induce platelet surface exposure of phosphatidylserine (PS) (Supplemental Figure 3D). Interestingly, platelets from DTA mice were hyperreactive in response to thrombin (Supplemental Figure 3B). This suggests that MSCs may act on the thrombin-mediated Par1/Par4 activation pathway and not the collagen-induced GPVI/GPIbα pathways or ADP-mediated pathways through P2Y1/P2Y12. Collectively, these findings suggest that stromal cells play a role in platelet production, assisting to lower the basal activation state of produced platelets and helping to maintain a normal ability to activate in response to thrombin.
Figure 1. Lepr+ stromal cells in the mouse BM help to maintain normal platelet numbers in the circulation and assist in lowering their basal activation levels.
Triple transgenic Lepr+tdTomato+DTA+ mice were generated. The efficiency of Lepr+ cell depletion was evaluated by (A) quantifying the remaining Lepr+ cells in the BM (n = 3–4). (B) We did not observe any significant difference in the number of megakaryocytes present in the femur of the DTA group compared with the control group (n = 6). (C) The number of platelets in the peripheral blood were quantified with an Advia coulter counter (n = 6). (D) Using flow cytometry, we detected a significant increase in the CD62P+ platelet activation level in the DTA group compared with the control at baseline (n = 4). (E) Representative histograms of CD62P+ platelet populations. (F) Fold change of CD62P+ platelet population relative to the control group (n = 4). (G) In vivo platelet clearance analysis of NHS-Biotin–labeled platelets over 6 days (n = 3–4). **P ≤ 0.01; ****P ≤ 0.0001. Two-tailed t tests were performed for A–G.
Isolation and characterization of UC tissue stromal cells.
We next tested whether stromal cells from human tissues have the same effect on platelet formation. Since cord blood–derived CD34 cells have been shown to differentiate into megakaryocytes with high efficiency compared with those isolated from peripheral blood and BM, we focused on identifying a compatible stromal cell from human UC tissue (23). Previous studies have identified MSC-like cells in human UC tissue, but the cells were not well defined (16–21). Unlike previously isolated UC tissue stromal cells, which have been primarily derived from Wharton’s jelly (17–19, 21), we focused on isolating arterial-derived stromal cells since perivascular cells around arteries have been shown to regulate hematopoietic cell production during development (24). Using flow cytometry, we found a population of CD51+PDGFRα+ arterial-derived stromal cells, termed cord-tissue Mesenchymal Stromal Cells (cMSCs), which were nonerythroid (CD235a–), nonendothelial (CD31–), and nonhematopoietic (CD45–). Of these stromal cells, 8.57% were CD51+ and PDGFRα+, 57.8% were CD51+ and PDGFRα–, and 32.7% were CD51– and PDGFRα– (Figure 2A). To confirm the location of these cells in the umbilical artery, we imaged UC tissue sections with confocal microscopy. Stromal cells positive for both CD51 and PDGFRα were located in the perivascular region (Figure 2B), whereas CD51+PDGFRα– cells were present within in the tunica media (Figure 2B).
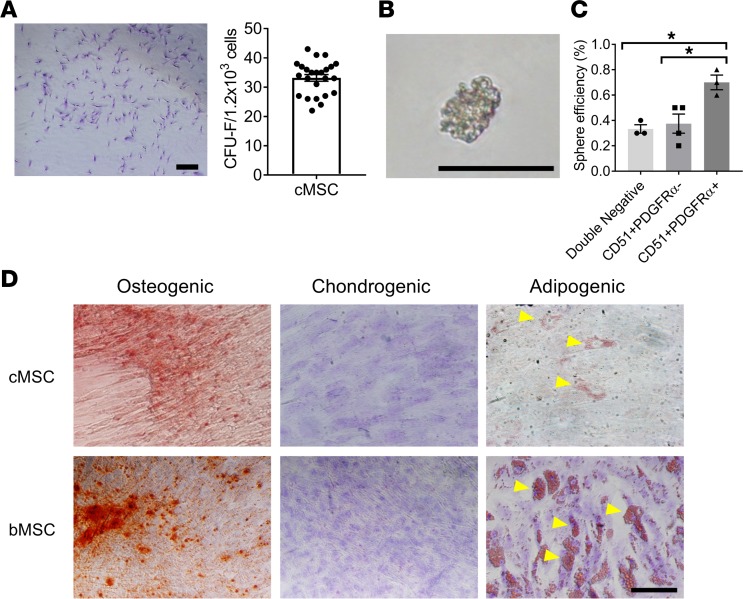
Figure 2. UC tissue stromal cell characterization.
(A) Using flow cytometry, we identified a population of stromal cells from the umbilical cord arteries that were CD45–, Ter119–, and CD31–. Of this population, approximately 57.8% were CD51+ and PDGFRα–, 8.57% were CD51+ and PDGFRα+, and 32.7% were CD51– and PDGFRα –. (B) Immunofluorescent-stained umbilical cord sections for CD31, CD51, and PDGFRα revealed a perivascular location around the umbilical artery for CD51+ and PDGFRα+ cells (marked by yellow arrowhead). Scale bar: 50 μm. (C) Using flow cytometry, CD51+PDGFRα+ cells were positive for various cell surface markers associated with MSCs (CD105, CD73, CD90, Stro-1, CD44, CD271, CD146, and PDGFRβ) and early embryonic cells (SSEA4 and GD2). (n = 3–4). (D) cMSCs expressed Nestin as shown by real-time PCR (n = 3–4). (E) cMSCs were enriched for a number of HSC maintenance genes, similar to BM MSCs (n = 4). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Two-tailed t tests were performed.
To further elucidate the MSC phenotype among the UC-derived stromal cell population, we investigated their cell surface protein expression and genomic profile. These cells expressed a panel of surface markers representative of MSCs (25), including CD105, CD90, CD73, CD271, CD44, CD146, PDGFRβ, and Stro-1, as well as early embryonic cell markers, including SSEA4 and GD2 (Figure 2C). Additionally, the cMSCs were positive for Nestin gene expression (Figure 2D and Supplemental Table 1), although the expression was lower than that of bMSCs (Figure 2D). Furthermore, cMSCs were enriched for a variety of other HSC maintenance genes, similar to bMSCs (Figure 2E).
When plated at a clonal density, cMSCs were able to adhere and propagate as individual colonies (Figure 3A), which is an important feature of MSCs. To assess their ability to self-renew in culture, cMSCs were subjected to a spheroid formation assay (12, 26). After 1 week in culture, cMSCs were able to divide and propagate as nonadherent cell spheroids (Figure 3B). The CD51+PDGFRα+ population of cMSCs was able to self-renew and produce spheroids at a significantly higher efficiency (0.7%) compared with the CD51+PDGFRα– (0.38%) population and the CD51–PDGFRα– (0.33%) population (Figure 3C). Lastly, cMSCs were investigated for their potential to differentiate from a stem cell state to restricted lineages. cMSCs cultured as a monolayer were able to undergo osteogenic (Figure 3D), chondrogenic, and adipogenic differentiation, similar to human bMSCs (Figure 3D). However, the level of adipogenic differentiation was lower in UC tissue stromal cells compared with those from BM (Figure 3D). Collectively, the genetic expression, cell-surface expression, self-renewal capacity, and ability to undergo trilineage differentiation suggest that these UC-derived cells are enriched for MSC activity and share a similar phenotype with those found in BM.
Figure 3. cMSC stem cell properties.
(A) cMSCs were able to adhere and produce colonies as demonstrated with Giemsa staining. Scale bar: 2 mm. The number of CFU-fibroblasts (CFU-F) were quantified per 1.2 × 103 seeded cells (n = 24). (B) cMSCs were able to self-renew and form mesenspheres in vitro. Scale bar: 1 mm. (C) Quantification of the sphere-forming ability of the CD51+PDGFRα+ population compared with the CD51+ PDGFRα– and CD51– PDGFRα– populations (n = 5–6). (D) cMSCs were able to undergo trilineage differentiation into the osteogenic lineage, as shown with Alizarin red staining, chondrogenic lineage as shown with Toluidine blue staining, and adipogenic lineage as shown with Oil Red O staining (top row), similar to human BM MSCs (bMSCs) (bottom row). Scale bar: 1 mm. *P ≤ 0.05. One-way ANOVA with Tukey post hoc test was performed. Yellow arrows indicate lipid droplets.
cMSCs enhance platelet formation in vitro.
Megakaryocyte induction from cord blood–derived CD34+ cells was conducted using previously established methods (4). Following an 8-day differentiation protocol, flow cytometry analysis revealed a decrease in CD34 expression (Figure 4A) coinciding with an increase in mature megakaryocytes expressing both CD41a and CD42b (Figure 4B), demonstrating the differentiation of hematopoietic progenitor cells into the megakaryocyte lineage.
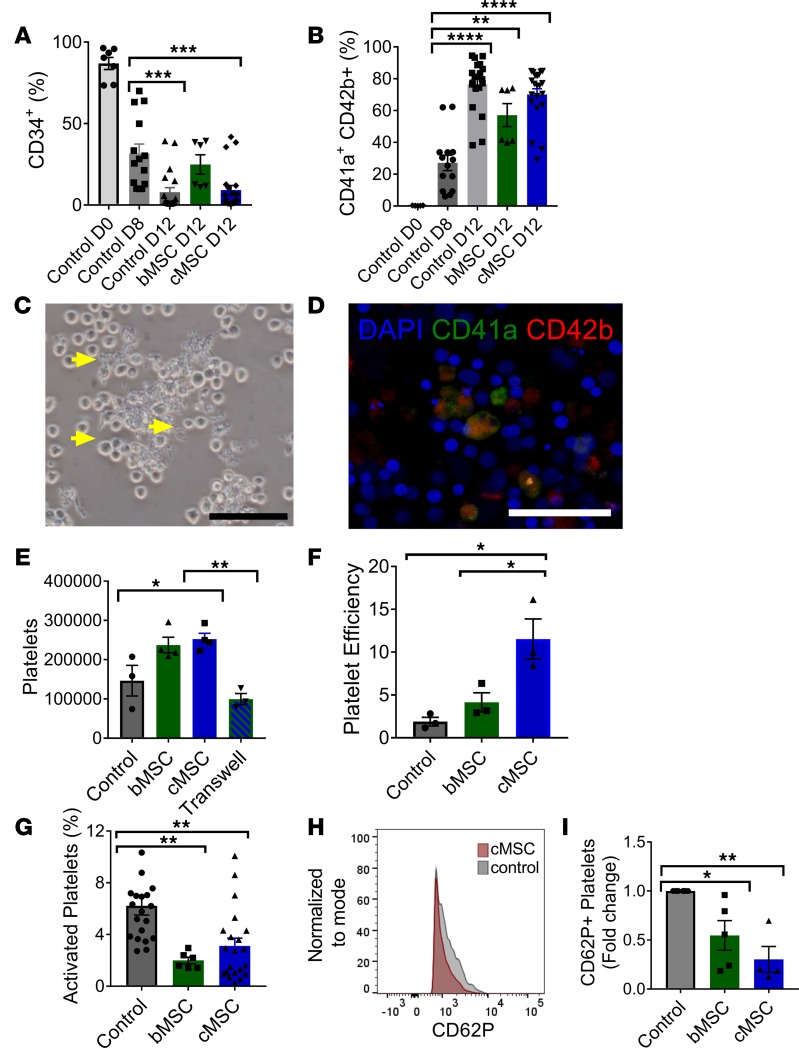
Figure 4. cMSCs cultured as a monolayer enhance platelet formation in vitro.
CD34+ cells were differentiated for 8 days into megakaryocytes and then cultured with cMSCs, with bMSCs, or alone. (A) Expression of CD34 decreased by day 8 of culture, demonstrating differentiation from the stem cell state. (B) In all 3 groups, the presence of CD41a+ and CD42b+ cells, representative of mature megakaryocytes, could be detected by flow cytometry. (C) By day 12 of in vitro culture, proplatelet formation could be detected in the cMSC coculture group. Scale bar: 50 μm. Yellow arrows indicate proplatelets. (D) Immunofluorescent staining of the cMSC coculture group at day 12 of culture revealed the presence of CD41a+CD42b+ megakaryocytes. Scale bar: 50 μm. (E) By flow cytometry, the number of platelets formed was quantified per culture well. Significantly fewer platelets were formed when cMSCs were separated by a transwell, suggesting that contact is required between megakaryocytes and cMSCs. (F) The platelet efficiency, or platelets formed per megakaryocyte, was significantly higher in the cMSC group compared with the control group. (G) The amount of activated platelets expressing CD62P was quantified by flow cytometry. (H) Representative histogram of CD62P+ platelet population in cMSC group versus control. (I) Fold change of CD62P+ platelets relative to the control group (n = 10–15 for all cocultures). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. All quantitative data was analyzed by 1-way ANOVA with Tukey post hoc test.
To determine if cMSCs can enhance platelet production from megakaryocytes, cMSCs were cultured as a monolayer with megakaryocytes differentiated for 8 days from CD34+ cells. After 4 days of coculture, proplatelets were seen extending from the megakaryocytes in culture with cMSCs (Figure 4C). As shown by immunofluorescence, these cells expressed mature megakaryocyte markers CD41a and CD42b (Figure 4D). We observed an increase (1.72-fold) in the number of platelets formed in the cMSC coculture group compared with the control group containing only megakaryocytes (Figure 4E).
Interestingly, megakaryocytes cocultured with cMSCs were more efficient at producing platelets (12 per input megakaryocyte) compared with megakaryocytes cocultured with bMSCs (4 per input megakaryocyte) and megakaryocytes cultured alone (2 per input megakaryocyte) (Figure 4F). Platelets generated in culture were examined for their basal activation level. Significantly fewer platelets in the coculture group expressed CD62P compared with the control group (Figure 4, G–I), suggesting that cMSCs may act to reduce platelet activation.
Since thrombopoietin (TPO) is key factor in promoting megakaryocyte maturation, we measured levels of TPO in our cultures. Increased TPO levels were detected in the cocultures containing cMSCs compared with the control group (Supplemental Figure 4, A and B). Analysis of cMSCs with real-time PCR revealed gene expression of THPO (Supplemental Figure 4C), suggesting that these cells are producing and secreting TPO in culture and corroborating other findings that suggest that stromal cells produce TPO (27, 28). We also found an increase in ploidy in the MSC coculture group compared with the control (Supplemental Figure 5), consistent with increased megakaryocyte maturation. We next investigated whether contact is required between the cMSCs and megakaryocytes to enhance platelet formation. Using a 0.4 μM transwell system, we seeded cMSCs in the upper chamber and cultured the megakaryocytes below. Interestingly, we did not observe an increase in platelet number after 4 days of coculture in the transwell system, suggesting that direct contact was required between cMSCs and megakaryocytes to promote improved platelet formation (Figure 4E).
We also evaluated whether cMSCs cultured as spheroids, as opposed to a monolayer, were able to enhance platelet formation from megakaryocytes differentiated from CD34+ cells. Spheroids cultured for 4 days in direct contact with megakaryocytes (Supplemental Figure 6A) were able to significantly increase the number of generated platelets compared with the control group (Supplemental Figure 6B). Similar to what we observed with the monolayer culture conditions, there was a decrease in the CD62P expression in the cultured platelets (Supplemental Figure 6C), as well as an increase in the number of platelets produced per input megakaryocyte progenitor cell (Supplemental Figure 6D). Interestingly, we observed an increase in the platelet efficiency in the spheroid-based cMSC coculture system (26 per input megakaryocyte; Supplemental Figure 6D) compared with the monolayer coculture system (12 per input megakaryocyte; Figure 4F).
Altogether, these data indicate that MSCs, through cell-to-cell contact with megakaryocytes, increase platelet formation both in monolayer form and even more so in spheroid form. More importantly, the platelets produced in this culture system are lower in activation state, an important consideration for developing a transfusion product.
Platelets produced by cMSC-stimulated megakaryocytes are functional.
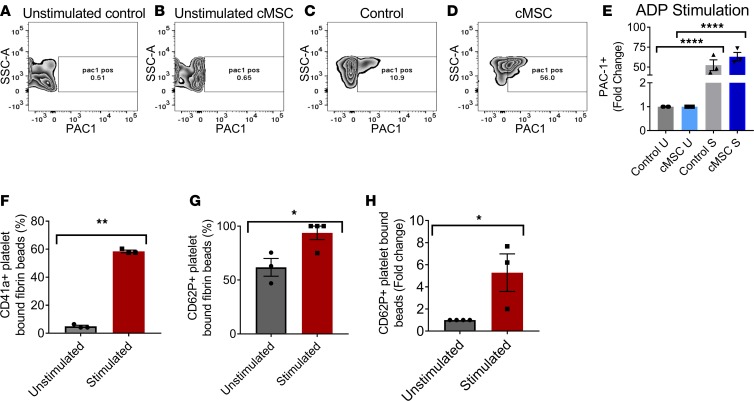
A series of functional tests were conducted on the collected platelets from the coculture system. Vital platelets were assessed by uptake of CalceinAM dye and evaluated by flow cytometry, gating on particles of human platelet size. At baseline, cMSC cocultured platelets displayed increased expression of GPIbα (Supplemental Figure 7A), suggesting that they are more mature. At baseline, both platelets from the cMSC and control group expressed very low levels of integrin αIIβ3 (Supplemental Figure 7B) and were not activated. The ability of platelets to activate in response to ADP agonist stimulation (200 μM) was assessed based on increased expression of PAC-1, measured by flow cytometry. After ADP agonist stimulation, generated platelets from the coculture system were able to significantly upregulate PAC-1 expression compared with the unstimulated group, confirming their functionality (Figure 5, A–E). Similarly, cMSC cocultured platelets were able to activate in response to thrombin stimulation (0.1 U/ml and 1 U/ml; Supplemental Figure 7C) and collagen (20 μg/ml and 80 μg/ml; Supplemental Figure 7D).
Figure 5. Functional platelet characterization, showing activation in response to ADP stimulation.
(A–D) Flow cytometry plots of PAC-1 expression in (A) unstimulated platelets from the control group and (B) unstimulated platelets from the cMSC group compared with (C) stimulated platelets from the control group and (D) stimulated platelets from the cMSC coculture group. (E) Quantification of the fold change in PAC-1 expression among the various groups (n = 3). A quantitative flow cytometry assay was performed, measuring the ability of platelets from the cMSC coculture group to adhere to matrix proteins bound to beads. (F) Platelets stimulated with ADP were able to bind efficiently to matrix-coated beads (n = 3–4). (G) Bound stimulated platelets were activated and expressed CD62P to a greater extent than unstimulated bound platelets (n = 3–4). (H) Fold change of stimulated CD62P+ platelets bound to matrix-coated beads relative to unstimulated platelets (n = 3–4). *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001. E was analyzed by 1-way ANOVA with Tukey post hoc test. F–H was analyzed by 2-tailed t tests.
We also assessed the capacity for platelets to adhere to damaged blood vessels using a quantitative flow cytometry assay with fibrinogen-coated microbeads to mimic the vessel extracellular matrix (29). Platelets from our coculture system were able to adhere to matrix-coated microbeads and increase their CD62P activation levels, as detected by flow cytometry (Figure 5, F–H).
The ability of platelets to spread on activated glass and remodel their cytoskeleton to form membrane protrusions indicative of early aggregation was observed with confocal microscopy. Platelets were stained with actin to confirm the presence of stress fibers due to cytoskeletal reorganization (Figure 6, A–F). When platelets were placed on a matrix-coated surface, they underwent further matrix reorganization and spreading (Figure 6, G–L), forming filopodia and lamellipodia, similar to primary donor platelets, demonstrating intact functionality.
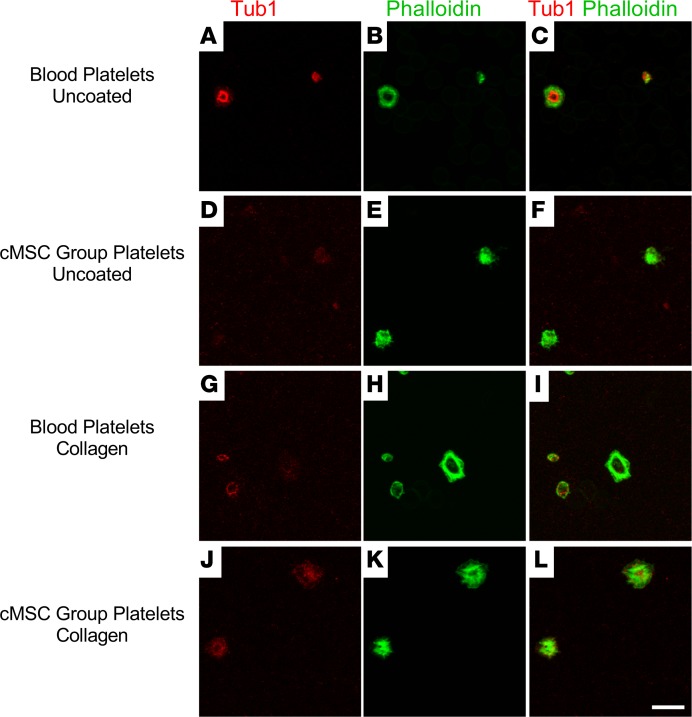
Figure 6. Generated platelets are able to spread on glass and matrix proteins.
(A–F) Platelets generated from cMSC cultures were positively stained with immunofluorescence for β1 tubulin and phalloidin (A–C), similar to native platelets from peripheral blood (D–F). (G–L) Platelets from the coculture group were able to spread on a collagen-coated surface (G–I), similar to native platelets (J–L). Scale bar: 10 μm.
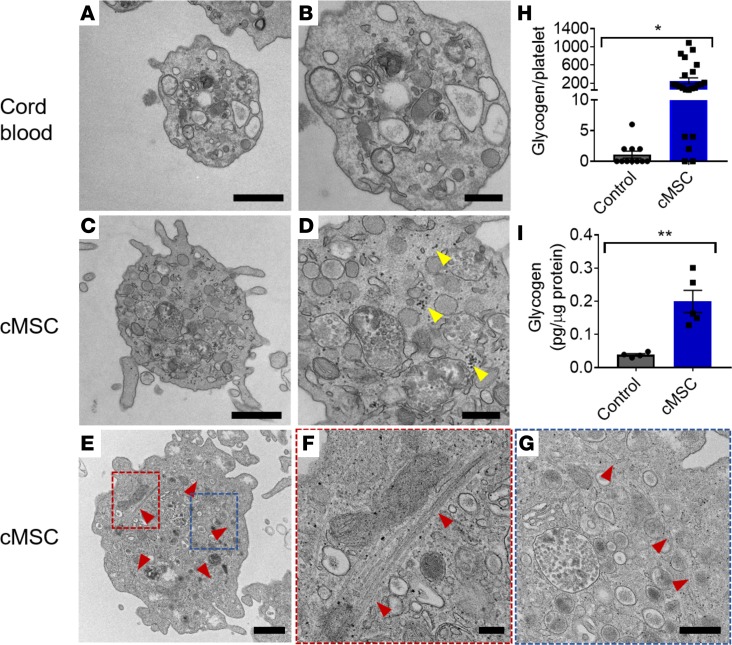
Coculture-expanded platelets were analyzed with transmission electron microscopy (TEM) imaging to compare their morphology and ultrastructural features with those of platelets from human cord blood. Similar to native platelets (Figure 7, A and B), platelets from the coculture group (Figure 7, C–G) displayed the presence of an open canalicular system, mitochondria, and α and dense granules. We also observed intact microtubule coils among our platelets (Figure 7, E–G), further demonstrating that the cMSC coculture system is able to maintain platelets at a resting state. Interestingly, there was a significant increase in the presence of glycogen in the cocultured platelets compared with donor platelets (Figure 7, H and I), suggesting that these platelets have unused energy reservoirs and are less metabolically active at their basal state.
Figure 7. Generated platelets have a similar ultrastructure to primary platelets.
(A–G) Representative transmission electron microscopy images of platelets from control cord blood platelets (A and B) and cMSC coculture group (C–G). Scale bars: 500 nm (A and B) and 200 nm (C and D). cMSC cocultured platelets contained α and dense granulates, glycogen sources (marked by yellow arrowheads), and mitochondria similar to platelets from the control group. (E–G) cMSC coculture platelet containing an intact microtubule coil at resting state. Red and blue regions are magnified in F and G, respectively. (H) Quantification of the number of glycogen granules per platelet section in the cMSC coculture group compared with cord blood platelets (n = 11–24). (I) ELISA assay quantification of glycogen content in platelets normalized to the protein content per cell (n = 4–5) comparing platelets from cMSC cocultures with platelets from megakaryocytes cultured alone. *P ≤ 0.05; **P ≤ 0.01. Two-tailed t tests were performed.
cMSCs promote megakaryocyte maturation and proplatelet formation, and they prevent platelet activation.
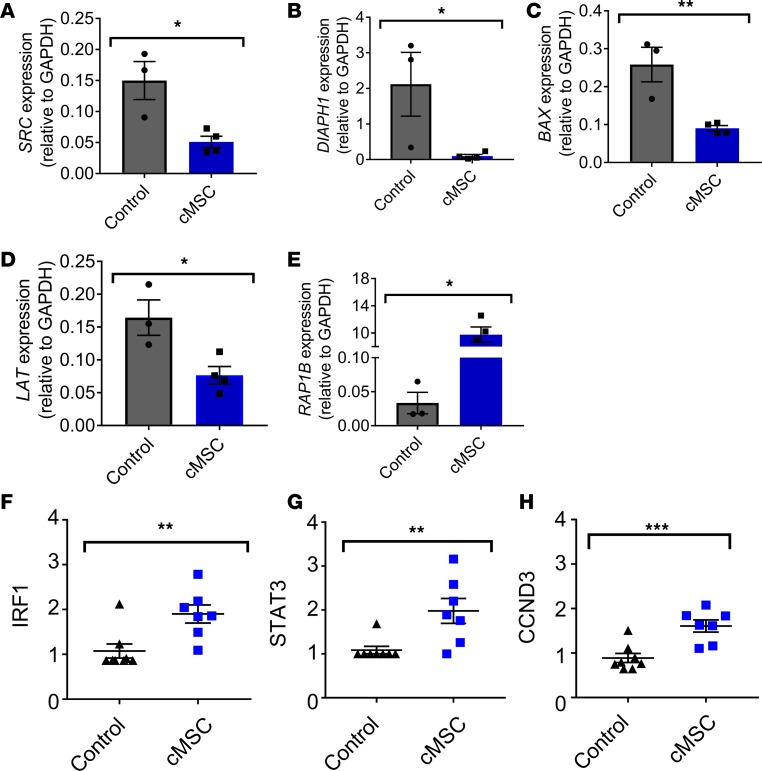
We next evaluated the expression of a number of genes known to play a role in increasing the potency of megakaryocytes to produce platelets. Src kinase inhibitors have been shown to increase megakaryocyte ploidy and in vitro platelet shedding (30). Furthermore, they have been known to increase platelet yield in CD34-derived megakaryocytes cultured in vitro (30). Interestingly, megakaryocytes cocultured with MSCs demonstrated lower expression of SRC, as analyzed by real-time PCR, compared with megakaryocytes cultured alone (Figure 8A and Supplemental Table 1). Another gene, DIAPH1, negatively regulates proplatelet formation by controlling the dynamics of actin and microtubule cytoskeleton production (31). Consistent with regulating proplatelet formation, we found that coculture with cMSCs decreased the expression of DIAPH1 compared with the control group (Figure 8B). Lastly, we observed a decreased expression of BAX, known to promote proplatelet formation and shedding (32) when downregulated, among the cMSC cocultured megakaryocytes compared with the control (Figure 8C).
Figure 8. cMSCs assist in promoting proplatelet formation and decrease platelet activation.
(A–C) Megakaryocytes from cMSC coculture group had decreased gene expression, as measured by real-time PCR, for SRC (A), DIAPH1 (B), and BAX (C), which are associated with increased megakaryocyte maturation (n = 3–4). (D and E) Furthermore, megakaryocytes from cMSC coculture group had decreased gene expression for LAT1 (D) and enriched for expression of RAP1B (E) relative to platelets from the control cultures, suggesting that cMSCs help to keep megakaryocytes in a lower activation state (n = 3–4). (F–H) Platelets from cMSC coculture group had increased protein expression, as measured by reverse phase protein analysis (RPPA), for IRF1 (F), STAT3 (G), and CCND3 (H), relative to platelets from the control cultures, suggesting that cMSCs are able to promote megakaryocyte maturation (n = 7–8). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Two-tailed t tests were performed.
We also assessed the expression of genes associated with lowering the basal activation state of formed platelets in megakaryocytes cocultured with cMSCs. Linker for Activation of T cells (LAT) has previously been shown to play a role in platelet activation (33). We observed significantly decreased expression of LAT in the coculture group compared with the control group (Figure 8D), corresponding to a decrease in CD62P expression (Figure 4, G–I, and Supplemental Figure 6C). Another gene, RAP1b, was shown in mouse models to be necessary for integrin-mediated platelet activation and efficient platelet aggregation (34). We observed an increased expression of RAP1B in the cMSC group compared with the control group (Figure 8E), suggesting that cMSCs may help to promote normal platelet aggregation.
To evaluate if the differentially expressed genes translated to changes in protein production, we conducted a reverse phase protein array (RPPA) comparing cMSC cocultured megakaryocytes with the control group (Supplemental Table 2). Interestingly, we observed an increase in IRF1 (Figure 8F) and STAT3 (Figure 8G) protein expression in the coculture group compared with the control, which has been shown to promote megakaryocyte differentiation and maturation mediated through TPO signaling, further supporting our observed increased TPO levels in the cocultures containing cMSCs compared with the control group (Supplemental Figure 4 and ref. 35). Finally, overexpression of Cyclin D3 (CCND3) has been shown to increase megakaryocyte number and ploidy (36). We observed an increase in CCND3 protein in our cMSC cocultures compared with the control (Figure 8H), along with an increase in ploidy (Supplemental Figure 5), further suggesting that cMSCs may assist in promoting megakaryocyte maturation.
Discussion
The ability to harness the innate function of coexisting cells in the megakaryocyte microenvironment may provide a novel model for improving platelet production in vitro. Human UC endothelial cells have been shown to enhance platelet production in an ex-vivo bioreactor setting (7). Additionally, human bMSCs have also been shown to improve platelet production, but the population of interest was poorly defined (15), leading to reproducibility challenges. In this potentially novel proof-of-principle study, we identify a population of CD51+PDGFRα+ perivascular stem cells found around the umbilical artery, which were rigorously characterized, and displayed a MSC phenotype. Remarkably, these cells were able to assist megakaryocytes to produce platelets in vitro, with a size and morphology similar to native human platelets. Further evaluation of the generated platelets revealed an ultrastructure resembling primary platelets, the ability to adhere and spread on matrix proteins, and the capacity to activate in response to agonist stimulation.
Newly formed platelets in vivo maintain low steady-state activation levels, even though they are subjected to high shear stress levels and apparent local turbulent forces (8, 37). A critical component in developing a successful platelet product is ensuring a low level of baseline activation. Deletion of perisinusoidal stromal cells in mice resulted in decreased circulating platelets with an increased activation state and hyperreactivity to stimulation by thrombin. Remarkably, in our culture system, cMSCs from human UC tissue similarly promoted low basal activation levels of platelets produced in vitro. Furthermore, we observed a decrease in LAT gene expression and an increase in RAP1B gene expression in the cMSC group compared with the control group, further supporting the appearance of low levels of platelet activation. TEM imaging of the platelet ultrastructure from the cMSC group further supported these findings, suggesting that these platelets have unused energy reservoirs and are less metabolically active at their basal state. Through TEM imaging and immunofluorescence, we demonstrate the presence of a microtubule coil around cMSC cocultured platelets, further suggesting that the platelets are maintaining a resting state before activation.
UC tissue is normally discarded after birth and represents an ideal tissue source for isolating MSCs. Whether cMSCs from UC tissue are comparable with other identified MSC populations from fetal and human adult tissues remains a topic for further study and may lead to additional sources of MSCs for enhancing platelet formation. Purified HSCs from UC blood units are already approved as a medical product by the US FDA, facilitating future translation of this technology. In our studies, we combined cMSCs and cord blood–derived HSCs from different patients — a process that was successfully able to improve platelet yields and may further simplify translation to the clinic.
In addition to generating platelets to serve as a new source for cellular therapy, this concept could possibly lead to the development of novel drugs for enhancing platelet production in patients with low platelet counts. Therapeutics targeting stromal cell populations in BM could potentially augment platelet production in patients with thrombocytopenia or hematological cancers. Conversely, the development of approaches to target stromal cells in order to lower platelet activation may reduce vascular inflammation or arterial thrombosis and directly benefit patients with cardiac disease.
Methods
Mouse experiments.
Mouse strains were obtained from The Jackson Laboratory and include B6.129(Cg)-Leprtm2(cre)Rck/J, B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, and B6.129P2-Gt(ROSA)26Sortm1(DTA)Lky/J. Lepr+DTA+ mice (for murine activation studies) were generated by crossing Lepr+ mice to DTA mice. Control mice contained Lepr but lacked DTA. Triple transgenic mice were generated by crossing Lepr+ mice to tdTomato mice, and the progeny were crossed with DTA mice. Control mice contained Lepr and tdTomato expression but lacked DTA.
Cell isolation from human UC tissue, cord blood, peripheral blood, and BM.
CD34+ cell isolation was conducted using previously established methods (4). Cord blood units were obtained from the National Cord Blood Program (Long Island City, New York, USA). To isolate CD34+ cells, whole blood was separated on a ficoll gradient (GE Healthcare) with Sepmate PBMC isolation tubes (Stemcell Technologies), and RBCs were lysed using Pharm Lyse (BD Biosciences). Magnetic beads bound to CD34 (Miltenyi Biotec) were used for positive cell selection. Cell phenotype was confirmed with flow cytometry prior to plating for cell culture. CD34+ cells were cultured until day 8 of megakaryocyte differentiation in media, similar to our previous publications, consisting of Stemspan SFEM II (Stemcell Technologies) supplemented with 50 ng/ml TPO (R&D Systems), 5 ng/ml SCF (R&D Systems), 1% penicillin/streptomycin (Invitrogen), and 1% nonessential amino acids (Invitrogen) (38, 39). Cells differentiated into megakaryocytes were cultured in media containing TPO only.
Donor UC tissue was received from the National Disease Research Interchange (NDRI; Philadelphia, Pennsylvania, USA). Fresh harvested UC tissue was sent to NYBC and processed within 24 hours of collection. Umbilical arteries were dissected and cut into 0.5-cm segments. To obtain a single cell suspension for subsequent cell sorting or flow cytometry analysis, artery segments were sequentially digested for 1.5 hours with PBS containing collagenase type 1 (562 U/ml; Invitrogen) and hyaluronidase (1 mg/ml; Thermo Fisher Scientific) and were supplemented with 1 M CaCl2 solution (MilliporeSigma) (40). Prior to cell culture, plates were coated with fibronectin (20 μg/ml; Stemcell Technologies) for 1 hour. Umbilical artery fragments were washed with PBS and placed on top of coated plates to allow cells to migrate and adhere to the plate. After 72 hours, artery segments were removed. Culture media for UC or BM stromal cells consisted of MEM-α (Invitrogen) supplemented with 1% penicillin/streptomycin (Invitrogen), 20% FBS (HyClone), and 10% Human MSC Supplement (Stemcell Technologies).
Human bMSCs were obtained from Lonza. These cells were confirmed positive for surface expression of CD29, CD44, CD105, CD166, CD90, and CD73 and were negative for CD14, CD34, CD45, HLA-DR, and CD19. Cells were seeded in 12-well plates and used for experiments at passage 3–5.
To isolate platelets from human peripheral blood or cord blood, blood was spun down at 300 g for 10 minutes at room temperature (acceleration 0, deceleration 0). For nonactivation assays, the platelet-rich plasma (PRP) was treated with 1 μl/ml of Prostaglandin E1 (MilliporeSigma; P5515-1MG) for 10 minutes at 37°C. PRP was transferred to a new polypropylene tube and further centrifuged at 2,500 g for 15 minutes at room temperature (acceleration 0, deceleration 0) to obtain the platelet pellet, which was then resuspended in HBSS (1×; Thermo Fisher Scientific, 14175-079).
To isolate platelets from mouse peripheral blood, mice were anesthetized with 1.5%–2.5% Isoflurane (Piramal; NDC 66794-017-25) and bled by puncture of the submandibular vein. Collected blood was diluted into 500 μl HBSS buffer with 50 μl ACD solution. PRP was obtained by centrifugation at 300 g for 10 minutes at room temperature (acceleration 0, deceleration 0). The supernatant was transferred to a new tube and spun down at 2,500 g for 15 minutes at room temperature (acceleration 0, deceleration 0).
Immunofluorescence.
UC tissues were fixed in 4% paraformaldehyde for 2 hours, placed in sequential gradients of increasing sucrose concentration, and embedded in Tissue-Tek OCT Compound (Electron Microscopy Sciences). Frozen sections 6 μm in thickness were generated at the Laboratory of Comparative Pathology of Memorial Sloan Kettering Cancer Center (New York, New York, USA).
Cultured cells, primary cells, and tissue sections were washed with PBS, followed by fixation with 4% paraformaldehyde. Cells were blocked for 2 hours at room temperature with PBS containing 0.1% Tween X-100 and 20% normal goat serum (Thermo Fisher Scientific). Primary and secondary antibodies were diluted in PBS containing 0.1% Tween X-100 and 2% normal goat serum. Samples were imaged at 10×–20× magnification on the Zeiss LSM 780 Laser Scanning Confocal Microscope and analyzed using Zen Blue Software (Zeiss).
To investigate platelet adhesion to matrix proteins, 24-well glass-bottom culture plates were coated with collagen (25 μg/ml; MilliporeSigma) for 30 minutes. Platelets were allowed to adhere for 1 hour at 37°C prior to fixation and staining, as detailed above. The following antibodies were used: Alexa-fluor 488 Phalloidin (Invitrogen, A12379) and Alexa-fluor 568 goat anti-rabbit (Invitrogen, A-11011); anti-β1 tubulin (Abcam, AB179511); and CD51 Alexa-fluor 488 (BioLegend, 920010, clone P1F6), CD140a Phycoerythrin (BioLegend, 323506, clone 16A1), CD31 eFluor 450 (eBioscience, 48-0319-42, clone WM-5a), CD41 Alexa-fluor 488 (BioLegend, 303703, clone HIP8), and CD42b Phycoerythrin (BioLegend, 303906, clone HIP1).
CFU fibroblast (CFU-F) assays.
UC-derived stromal cells were sorted and seeded at a clonal density (2,000 cells/well) in 12-well adherent tissue culture plates. Cells were cultured with media consisting of MEM-α supplemented with 20% FBS (Hyclone), 10% human MesenCult Stimulatory Supplement (Stemcell Technologies), and 0.5% penicillin-streptomycin solution (Invitrogen). After 10 days, formed colonies were stained with Giemsa (Electron Microscopy Science) and quantified.
ELISA assays.
To assess secreted TPO from the cocultures, supernatant was analyzed using the Human Thrombopoietin Quantikine ELISA Kit (R&D Systems). Glycogen content from the cocultured platelets was assessed using the Human Glycogen Synthase I ELISA kit (Abcam).
Sphere formation assays.
UC-derived stromal cells were sorted and seeded at a clonal density (1,000–2,000 cells/well) in 35 mm ultra-low adherence tissue culture plates (Stemcell Technologies) and cultured with media previously described (13, 41). Half of the media was changed weekly. Generated spheres were quantified after 10 days.
Trilineage differentiation assays.
Methods for differentiation into chondrogenic, adipogenic, or osteogenic lineages was conducted as in our previous studies (24, 41, 42). Briefly, adherent monolayers of bMSCs or UC tissue stromal cells were cultured to 75% confluence. The cell culture media was subsequently replaced with either MesenCult-ACF Chondrogenic Differentiation Medium (Stemcell Technologies), MesenCult Osteogenic Differentiation Kit (Stemcell Technologies) media, or Mesencult Adipogenic Differentiation Media (Stemcell Technologies). Toluidine blue was used to assess for chondrogenic differentiation, and Alizarin Red staining was performed to evaluate osteogenic differentiation using our previous methods (24, 41, 42). For adipogenic differentiation, cells were stained with Oil Red O. Briefly, cells were fixed with formalin, washed with tap water, rinsed with isopropanol, and stained with Oil Red O solution for 15 minutes. After another rinse with isopropanol, cells were counterstained with hematoxylin and rinsed with distilled water.
Electron microscopy.
Platelets were spun into pellets and fixed using 4% paraformaldehyde (Electron Microscopy Sciences) and 2% glutaraldehyde (Electron Microscopy Sciences) in 0.1 M sodium cacodylate (Electron Microscopy Sciences) at pH 7.4. After washing in sodium cacodylate buffer, pH 7.4 (Electron Microscopy Sciences), the samples were postfixed with a solution containing 1% osmium tetroxide (Electron Microscopy Sciences) and 1.5% potassium ferrocynide (Electron Microscopy Sciences). To visualize microtubules, platelets were treated with Triton X-100 (MilliporeSigma) and 0.1% glutaraldehyde (Electron Microscopy Sciences) prior to fixation with 1% glutaraldehyde for 2 hours. Subsequently, the samples were washed with water and stained en bloc with 1% uranyl acetate (Electron Microscopy Sciences). The platelets were dehydrated in a graded series of ethanol solutions (Electron Microscopy Sciences) and infiltrated with Eponate 12 (Electron Microscopy Sciences) using a microwave (PELCO Biowave, Ted Pella). Sections were cut at 70 nm, mounted on copper grids, and stained with uranyl acetate (Electron Microscopy Sciences) and lead citrate (Electron Microscopy Sciences). Stained grids were imaged at 80 kV on a JEOL 100CX TEM with a XR41-C AMT digital imaging system (Advantage Microscopy Technology Corp.) or a Talos120C TEM (Thermo Fisher Scientific) with a Gatan (4k × 4k) OneView Camera.
Flow cytometry and cell sorting.
Multiparameter flow cytometry analysis of antibody-stained cell suspensions was performed using LSRFortessa (BD Biosciences) and analyzed using FlowJo Software (FlowJo LLC). Vital platelets were assessed by uptake of CalceinAM dye (Thermo Fisher Scientific) and evaluated by flow cytometry, gating on particles of human or murine platelet size using primary peripheral blood platelets as a comparison. To enrich for specific cell populations, cells were sorted using a Moflo Cell Sorter (Beckman Coulter).
To assess the ability of platelets to activate, platelets were stimulated with ADP (MilliporeSigma; 20 μM or 200 μM), collagen (Thermo Fisher Scientific; 20 μg/ml or 80 μg/ml), or thrombin (MilliporeSigma; 0.1 U/ml or 1 U/ml) in HBSS with 1 mM CaCl2 for 15 minutes at 37°C. Murine platelet activation was assessed with CD62P (Invitrogen; 17-0626-82, clone, Psel.K02.3) and JON/A (Emfret, M023-2), and human platelet activation was assessed using CD62P (BioLegend, 304918; clone AK4) and PAC-1 (BioLegend, 362806). To test the ability of platelets to adhere to matrix-coated beads, 20 μm polystyrene microbeads (Polyscience) were coated with fibrinogen (MilliporeSigma; 8 mg/ml) for 30 minutes. As a control, beads were coated with BSA (MilliporeSigma). To assess PS exposure among platelets, Annexin V-APC (BioLegend, 640912) expression was measured following collagen stimulation (80 μg/ml). Phospho-serine (MilliporeSigma, 05-1000x) and Phospho-tyrosine (Abcam, AB179530) levels in murine platelets were assessed at baseline using goat anti-mouse (Invitrogen, A32728) or goat anti-rabbit (Invitrogen, P10994) secondary antibodies, respectively. Intracellular vWF was measured to assess α-granule numbers (Abcam, AB11713) using donkey anti-sheep secondary antibody (Invitrogen, A21448).
For megakaryocyte ploidy analysis, cells surface stained with CD41a (eBioscience, 12-0419-42, clone HIP8), and CD41b antibodies (BioLegend, 354804, clone HIP2) were submerged in fixation/permeabilization solution (BD Biosciences) and stained with propidium iodide solution (2 μg/ml, Invitrogen).
Platelet clearance analysis.
Murine platelets were biotinylated by dissolving 10 mg/kg of Biotin-NHS (MilliporeSigma) in DMSO diluted into 200 μl of sterile PBS. The solution was injected i.v. through the retroorbital plexus. Blood was drawn from the mice 1 hour after injection to assess the baseline level of biotinylation by staining with streptavidin-phycoerythrin/Cy7 (BioLegend, 405206) and measuring by flow cytometry. Blood was collected every 24 hours for 6 days to assess the remaining biotinylated platelets.
Genomic and protein expression analysis.
For genomic analysis, collected cells were lysed and subjected to mRNA isolation using the Dynabeads mRNA Direct kit (Thermo Fisher Scientific). Reverse transcription to produce cDNA was conducted using RNA to cDNA Ecodry Premix (Takara). Quantitative PCR was performed using SYBR Green (Roche Diagnostics). Samples were analyzed using the ViiA 7 Real-Time PCR System (Applied Biosystems). Relative gene expression was calculated based on the comparative Ct method and normalized to the reference gene GAPDH. Primer sequences are listed in Supplemental Table 1.
RPPA analysis was conducted by the Functional Proteomics RPPA core facility at MD Anderson Cancer Center (Houston, Texas, USA) on protein lysates extracted from our cultured megakaryocytes. Cellular proteins were denatured by 1% SDS (with β-mercaptoethanol) and diluted in five 2-fold serial dilutions in lysis buffer. Lysates were arrayed on nitrocellulose-coated slides (Grace Bio Lab) by Aushon 2470 Arrayer (Aushon BioSystems). A total of 5,808 array spots were arranged on each slide, including the spots corresponding to serial diluted: (a) standard lysates or (b) positive and negative controls prepared from mixed cell lysates or dilution buffer. Each slide was probed with a validated primary antibody plus a biotin-conjugated secondary antibody. The signal obtained was amplified using a Dako Cytomation–Catalyzed system (Dako) and visualized by DAB colorimetric reaction. The slides were scanned, analyzed, and quantified using a customized software to generate spot intensity. Each dilution curve was fitted with a logistic model. The protein concentrations of each set of slides were then normalized for protein loading.
Statistics.
All statistical analysis was performed using Graphpad Prism 7. Statistical significance between 2 samples was determined using unpaired 2-tailed Student’s t tests. For multiple sample comparison, 1-way ANOVA was done using Tukey post hoc test for multiple comparisons correction or repeated measure correction. P <0.05 was considered statistically significant. All quantitative data is represented by mean ± SEM.
Study approval.
All human tissue specimens were designated exempt by the IRB of the NYBC. All animal experiments were approved by the IACUC of the NYBC.
Author contributions
AM designed, performed, and analyzed all of the experiments. WB assisted with coculture experiments and differentiation assays. PR was instrumental in developing the cell isolation protocols and conducting flow cytometry. GF and SO analyzed the RPPA studies and assisted with genomic analysis. ANS assisted with the in vivo and in vitro activation studies. HZ, XA, and CL provided input on experimental design, in addition to data analysis. KY helped with experimental design, input with data analysis, and overall project supervision. AM wrote the manuscript, while KY edited the manuscript with consultation and contribution from all authors.
Supplementary Material
Acknowledgments
We are grateful to Beau Mitchell for initial discussions and help with protocol development. We thank the National Cord Blood Program for providing cord blood for research use. The authors would like to thank the Mihaela Barbu-Stevanovic, Sean D’Italia, and Zannatul Monia for their guidance with flow cytometry and cell sorting. Additionally, we appreciate the help of Vijendra Ramlall (NYBC), Yunfeng Liu (NYBC), and David Shi (NYBC) for their technical assistance with experiments and data analysis. We would like to thank Denis Voronin (NYBC) and the Electron Microscopy Facility at Rockefeller University for their guidance with TEM imaging. We also thank NYU Langone Health DART Microscopy Lab Alice Liang, Chris Petzold, Joseph Sall, and Kristen Dancel-Manning for their assistance with TEM work. We would like to thank the Functional Proteomics RPPA Core facility at MD Anderson Cancer Center for assisting with the protein analysis studies. The Functional Proteomics RPPA Core facility is supported by MD Anderson Cancer Center Support Grant 5 P30 CA016672-40. The NYU Langone Health DART Microscopy Lab is partially funded by NYU Cancer Center Support Grant NIH/NCI P30CA016087. This work was also supported in part by funds from The National Blood Foundation Early Career Scientific Research Grants Program (AM), the Rose M. Badgeley Residuary Charitable Trust (AM), NIH R21HL139695 (AM), and NIH R01HL122788 (KY). We would like to thank Vijay Nandi (NYBC) for her assistance with statistical analysis.
Version 1. 08/22/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(16):e126982. https://doi.org/10.1172/jci.insight.126982.
Contributor Information
Avital Mendelson, Email: amendelson@nybc.org.
Weili Bao, Email: wbao@nybloodcenter.org.
Peter Rosston, Email: peter@rosston.com.
Georgia Fallon, Email: gf2341@columbia.edu.
Sophie Ohrn, Email: sohrn@nybc.org.
Hui Zhong, Email: hzhong@nybloodcenter.org.
Cheryl Lobo, Email: clobo@nybc.org.
Xiuli An, Email: XAn@nybc.org.
Karina Yazdanbakhsh, Email: kyazdanbakhsh@nybc.org.
References
- 1.Pang L, Weiss MJ, Poncz M. Megakaryocyte biology and related disorders. J Clin Invest. 2005;115(12):3332–3338. doi: 10.1172/JCI26720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machlus KR, Italiano JE. The incredible journey: From megakaryocyte development to platelet formation. J Cell Biol. 2013;201(6):785–796. doi: 10.1083/jcb.201304054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pulecio J, et al. Direct Conversion of Fibroblasts to Megakaryocyte Progenitors. Cell Rep. 2016;17(3):671–683. doi: 10.1016/j.celrep.2016.09.036. [DOI] [PubMed] [Google Scholar]
- 4.Avanzi MP, Oluwadara OE, Cushing MM, Mitchell ML, Fischer S, Mitchell WB. A novel bioreactor and culture method drives high yields of platelets from stem cells. Transfusion. 2016;56(1):170–178. doi: 10.1111/trf.13375. [DOI] [PubMed] [Google Scholar]
- 5.Feng Q, et al. Scalable generation of universal platelets from human induced pluripotent stem cells. Stem Cell Reports. 2014;3(5):817–831. doi: 10.1016/j.stemcr.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blin A, et al. Microfluidic model of the platelet-generating organ: beyond bone marrow biomimetics. Sci Rep. 2016;6:21700. doi: 10.1038/srep21700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thon JN, et al. Platelet bioreactor-on-a-chip. Blood. 2014;124(12):1857–1867. doi: 10.1182/blood-2014-05-574913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito Y, et al. Turbulence Activates Platelet Biogenesis to Enable Clinical Scale Ex Vivo Production. Cell. 2018;174(3):636–648.e18. doi: 10.1016/j.cell.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Asada N, et al. Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat Cell Biol. 2017;19(3):214–223. doi: 10.1038/ncb3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014;20(8):833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asada N, Takeishi S, Frenette PS. Complexity of bone marrow hematopoietic stem cell niche. Int J Hematol. 2017;106(1):45–54. doi: 10.1007/s12185-017-2262-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinho S, et al. PDGFRα and CD51 mark human nestin+ sphere-forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J Exp Med. 2013;210(7):1351–1367. doi: 10.1084/jem.20122252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Méndez-Ferrer S, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466(7308):829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avecilla ST, et al. Chemokine-mediated interaction of hematopoietic progenitors with the bone marrow vascular niche is required for thrombopoiesis. Nat Med. 2004;10(1):64–71. doi: 10.1038/nm973. [DOI] [PubMed] [Google Scholar]
- 15.Cheng L, Qasba P, Vanguri P, Thiede MA. Human mesenchymal stem cells support megakaryocyte and pro-platelet formation from CD34(+) hematopoietic progenitor cells. J Cell Physiol. 2000;184(1):58–69. doi: 10.1002/(SICI)1097-4652(200007)184:1<58::AID-JCP6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 16.Kandoi S, et al. Evaluation of platelet lysate as a substitute for FBS in explant and enzymatic isolation methods of human umbilical cord MSCs. Sci Rep. 2018;8(1):12439. doi: 10.1038/s41598-018-30772-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh JY, et al. Mesenchymal stem cells from human umbilical cord express preferentially secreted factors related to neuroprotection, neurogenesis, and angiogenesis. PLoS ONE. 2013;8(8):e72604. doi: 10.1371/journal.pone.0072604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goyal U, Ta M. p53-NF-κB Crosstalk in Febrile Temperature-Treated Human Umbilical Cord-Derived Mesenchymal Stem Cells. Stem Cells Dev. 2019;28(1):56–68. doi: 10.1089/scd.2018.0115. [DOI] [PubMed] [Google Scholar]
- 19.Badner A, Hacker J, Hong J, Mikhail M, Vawda R, Fehlings MG. Splenic involvement in umbilical cord matrix-derived mesenchymal stromal cell-mediated effects following traumatic spinal cord injury. J Neuroinflammation. 2018;15(1):219. doi: 10.1186/s12974-018-1243-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moroncini G, et al. Mesenchymal stromal cells from human umbilical cord prevent the development of lung fibrosis in immunocompetent mice. PLoS ONE. 2018;13(6):e0196048. doi: 10.1371/journal.pone.0196048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao D, et al. Hypoxia with Wharton’s jelly mesenchymal stem cell coculture maintains stemness of umbilical cord blood-derived CD34+ cells. Stem Cell Res Ther. 2018;9(1):158. doi: 10.1186/s13287-018-0902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger G, Hartwell DW, Wagner DD. P-Selectin and platelet clearance. Blood. 1998;92(11):4446–4452. [PubMed] [Google Scholar]
- 23.van den Oudenrijn S, von dem Borne AE, de Haas M. Differences in megakaryocyte expansion potential between CD34(+) stem cells derived from cord blood, peripheral blood, and bone marrow from adults and children. Exp Hematol. 2000;28(9):1054–1061. doi: 10.1016/S0301-472X(00)00517-8. [DOI] [PubMed] [Google Scholar]
- 24.Khan JA, et al. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science. 2016;351(6269):176–180. doi: 10.1126/science.aad0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv FJ, Tuan RS, Cheung KM, Leung VY. Concise review: the surface markers and identity of human mesenchymal stem cells. Stem Cells. 2014;32(6):1408–1419. doi: 10.1002/stem.1681. [DOI] [PubMed] [Google Scholar]
- 26.Isern J, et al. Self-renewing human bone marrow mesenspheres promote hematopoietic stem cell expansion. Cell Rep. 2013;3(5):1714–1724. doi: 10.1016/j.celrep.2013.03.041. [DOI] [PubMed] [Google Scholar]
- 27.Sungaran R, Chisholm OT, Markovic B, Khachigian LM, Tanaka Y, Chong BH. The role of platelet alpha-granular proteins in the regulation of thrombopoietin messenger RNA expression in human bone marrow stromal cells. Blood. 2000;95(10):3094–3101. [PubMed] [Google Scholar]
- 28.Guerriero A, Worford L, Holland HK, Guo GR, Sheehan K, Waller EK. Thrombopoietin is synthesized by bone marrow stromal cells. Blood. 1997;90(9):3444–3455. [PubMed] [Google Scholar]
- 29.Tynngård N, Wallstedt M, Södergren AL, Faxälv L, Ramström S. Platelet adhesion changes during storage studied with a novel method using flow cytometry and protein-coated beads. Platelets. 2015;26(2):177–185. doi: 10.3109/09537104.2014.891728. [DOI] [PubMed] [Google Scholar]
- 30.Jarocha D, Vo KK, Lyde RB, Hayes V, Camire RM, Poncz M. Enhancing functional platelet release in vivo from in vitro-grown megakaryocytes using small molecule inhibitors. Blood Adv. 2018;2(6):597–606. doi: 10.1182/bloodadvances.2017010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan J, et al. The formin DIAPH1 (mDia1) regulates megakaryocyte proplatelet formation by remodeling the actin and microtubule cytoskeletons. Blood. 2014;124(26):3967–3977. doi: 10.1182/blood-2013-12-544924. [DOI] [PubMed] [Google Scholar]
- 32.Josefsson EC, et al. Megakaryocytes possess a functional intrinsic apoptosis pathway that must be restrained to survive and produce platelets. J Exp Med. 2011;208(10):2017–2031. doi: 10.1084/jem.20110750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragab A, et al. Roles of the C-terminal tyrosine residues of LAT in GPVI-induced platelet activation: insights into the mechanism of PLC gamma 2 activation. Blood. 2007;110(7):2466–2474. doi: 10.1182/blood-2007-02-075432. [DOI] [PubMed] [Google Scholar]
- 34.Chrzanowska-Wodnicka M, Smyth SS, Schoenwaelder SM, Fischer TH, White GC. Rap1b is required for normal platelet function and hemostasis in mice. J Clin Invest. 2005;115(3):680–687. doi: 10.1172/JCI22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Z, Richmond TD, Muntean AG, Barber DL, Weiss MJ, Crispino JD. STAT1 promotes megakaryopoiesis downstream of GATA-1 in mice. J Clin Invest. 2007;117(12):3890–3899. doi: 10.1172/JCI33010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmet JM, Toselli P, Ravid K. Cyclin D3 and megakaryocyte development: exploration of a transgenic phenotype. Stem Cells. 1998;16 Suppl 2:97–106. doi: 10.1002/stem.5530160713. [DOI] [PubMed] [Google Scholar]
- 37.Junt T, et al. Dynamic visualization of thrombopoiesis within bone marrow. Science. 2007;317(5845):1767–1770. doi: 10.1126/science.1146304. [DOI] [PubMed] [Google Scholar]
- 38.Avanzi MP, Izak M, Oluwadara OE, Mitchell WB. Actin inhibition increases megakaryocyte proplatelet formation through an apoptosis-dependent mechanism. PLoS ONE. 2015;10(4):e0125057. doi: 10.1371/journal.pone.0125057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avanzi MP, Chen A, He W, Mitchell WB. Optimizing megakaryocyte polyploidization by targeting multiple pathways of cytokinesis. Transfusion. 2012;52(11):2406–2413. doi: 10.1111/j.1537-2995.2012.03711.x. [DOI] [PubMed] [Google Scholar]
- 40.Smith JR, Cromer A, Weiss ML. Human Umbilical Cord Mesenchymal Stromal Cell Isolation, Expansion, Cryopreservation, and Characterization. Curr Protoc Stem Cell Biol. 2017;41:1F.18.1–1F.18.23. doi: 10.1002/cpsc.24. [DOI] [PubMed] [Google Scholar]
- 41.Mizoguchi T, et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev Cell. 2014;29(3):340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendelson A, et al. Chondrogenesis by chemotactic homing of synovium, bone marrow, and adipose stem cells in vitro. FASEB J. 2011;25(10):3496–3504. doi: 10.1096/fj.10-176305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.