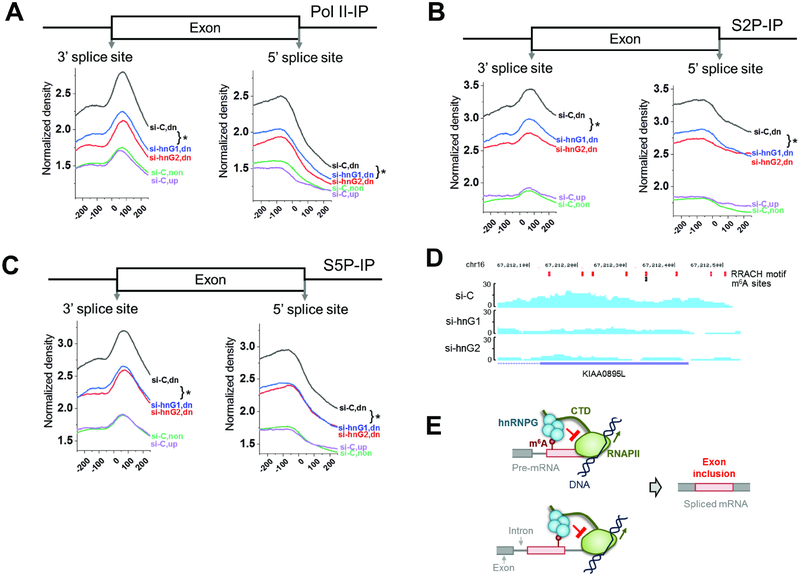
Figure 7: m6A-dependent regulation of alternative splicing by hnRNPG and RNAPII occupancy.
A. RNAPII ChIP-seq upon hnRNPG knockdown, using total RNAPII antibody. si-C: control knockdown; hnG1, hnG2: hnRNPG knockdown with two siRNAs. Co-regulated exons down- (dn, 15318), up-(up, 11255), and non-regulated (non, 35377) upon hnRNPG or METTL3/14 knockdown. * p < 10−4.
B. Same as (A) using S2P-CTD antibody.
C. Same as (A) using S5P-CTD antibody.
D. ChIP-seq tracks from KIAA0895L gene using RNAPII-S2P antibody in cells treated with control or hnRNPG siRNA. si-C: control siRNA; si-hnG1, si-hnG2: hnRNPG knockdown with two siRNAs. RRACH motifs: red; m6A sites: black.
E. Model for m6A-dependent regulation of exon inclusion by hnRNPG. Red circle: m6A site; red box: alternative exon; gray: constitutive exon; black lines: DNA; green: RNAPII with CTD as extended line; cyan: hnRNPG complex. RNAPII transcribes through the splice site, m6A is installed, and hnRNPG interacts with m6A, causing RNAPII to increase dwell time downstream of the m6A site, which results in hnRNPG- and m6A-dependent exon inclusion.