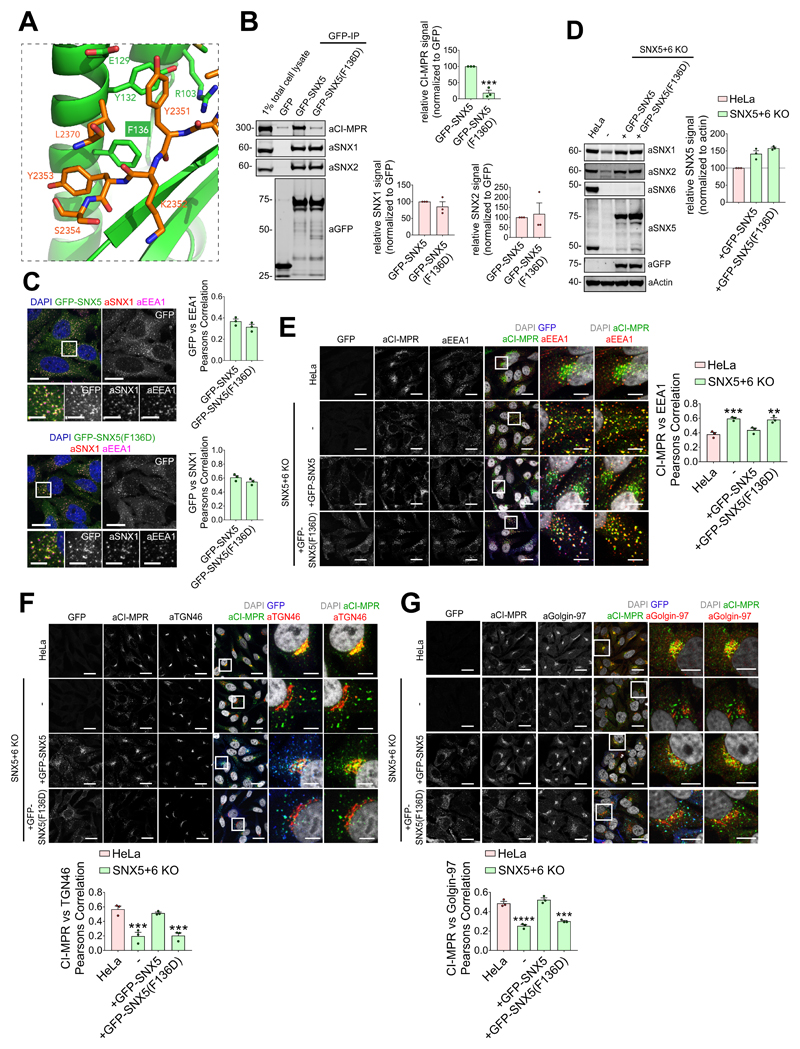
Figure 4. Interaction between CI-MPR and the hydrophobic groove of SNX5 PX domain is required for CI-MPR retrograde trafficking.
(A) CI-MPR peptide bound to SNX5 hydrophobic groove highlighting Phe136 position. (B) Co-immunoprecipitation of GFP-tagged SNX5 and SNX5(F136D) transiently transfected in HEK293T cells, summary of relative binding. Quantification of band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to GFP expression, presented as average fraction of GFP-SNX5 control. GFP-SNX5(F136D) mutant compared with GFP-SNX5 using two-tailed Unpaired t-test (***P<0.001; P values for SNX1: P=0.3655, SNX2: P=0.7646; CI-MPR: P=0.0006). (C) F136D mutation in SNX5 PX domain does not affect SNX5 endosomal localization. HeLa cells lentivirally transduced with GFP-SNX5 or GFP-SNX5(F136D) viral particles. Transduced cells fixed and stained for EEA1 and SNX1. Cell numbers analysed for colocalization between GFP and EEA1: GFP-SNX5: 50 cells; GFP-SNX5(F136D): 57 cells across n=3 independent experiments. Pearson’s coefficient values compared using two-tailed Unpaired t-test (P=0.1862). Cell numbers analysed for colocalization between GFP and SNX1: GFP-SNX5: 51 cells; GFP-SNX5(F136D): 59 cells across n=3 independent experiments. Pearson’s coefficient values compared using Unpaired t-test (P=0.1771). Scale bars 20 μm (non-zoom images) and 10 μm (insets). (D) GFP-SNX5 and GFP-SNX5(F136D) reintroduced in SNX5+SNX6 KO cells at levels comparable to endogenous. HeLa SNX5+SNX6 KO clonal line lentivirally transduced with GFP-SNX5 or GFP-SNX5(F136D) viral particles and SNX5-levels analyzed by quantitative fluorescence-based western blotting. Band intensities measured using Odyssey software and normalized to β-Actin before calculating percentage of protein compared with parental HeLa control. Bars represent mean of n=3 independent experiments. (E, F and G) Distribution of endogenous CI-MPR in HeLa cells, HeLa SNX5+SNX6 KO clonal line and HeLa SNX5+SNX6 KO clonal line transduced with GFP-SNX5 or GFP-SNX5(F136D). Colocalization analysis between CI-MPR and EEA1, CI-MPR and TGN46, and CI-MPR and Golgin-97. Cell numbers analysed across n=3 independent experiments for CI-MPR and EEA1 colocalisation (E): HeLa: 129 cells, SNX5+SNX6 KO: 144 cells, KO+GFP-SNX5: 159 cells; KO+GFP-SNX5(F136D): 122 cells. Pearson’s coefficient values compared with HeLa control using one-way ANOVA and Dunnett’s test (***P<0.001, **P<0.01; SNX5+SNX6 KO: P=0.0007, KO+GFP-SNX5: P=0.2886, KO+GFP-SNX5(F136D): P=0.0010). Cell numbers analysed across n=3 independent experiments for CI-MPR and TGN46 colocalisation (F): HeLa: 126 cells, SNX5+SNX6 KO: 111 cells, KO + GFP-SNX5: 121 cells; KO+GFP-SNX5(F136D): 110 cells. Pearson’s coefficient values compared with HeLa control using one-way ANOVA and Dunnett’s test (***P<0.001; SNX5+SNX6 KO: P=0.0005, KO+GFP-SNX5: P=0.6897, KO+GFP-SNX5(F136D): P=0.0006). Cell numbers analysed across n=3 independent experiments for CI-MPR and Golgin-97 colocalisation (G): HeLa: 105 cells, SNX5+SNX6 KO: 127 cells, KO+GFP-SNX5: 122 cells; KO+GFP-SNX5(F136D): 131 cells. Pearson’s coefficient values compared with HeLa control using one-way ANOVA and Dunnett’s test ****P<0.0001, ***P<0.001; SNX5+SNX6 KO: P=<0.0001, KO+GFP-SNX5: P=0.3482, KO+GFP-SNX5(F136D): P=0.0002). Scale bars 20 μm (micrographs) and 5 μm (insets). (B-D,G) Bars, mean; error bars, SEM; circles represent individual data points. Unprocessed original scans of immunoblots: Supplementary Figure 3. Statistics source data: Supplementary Table 4.