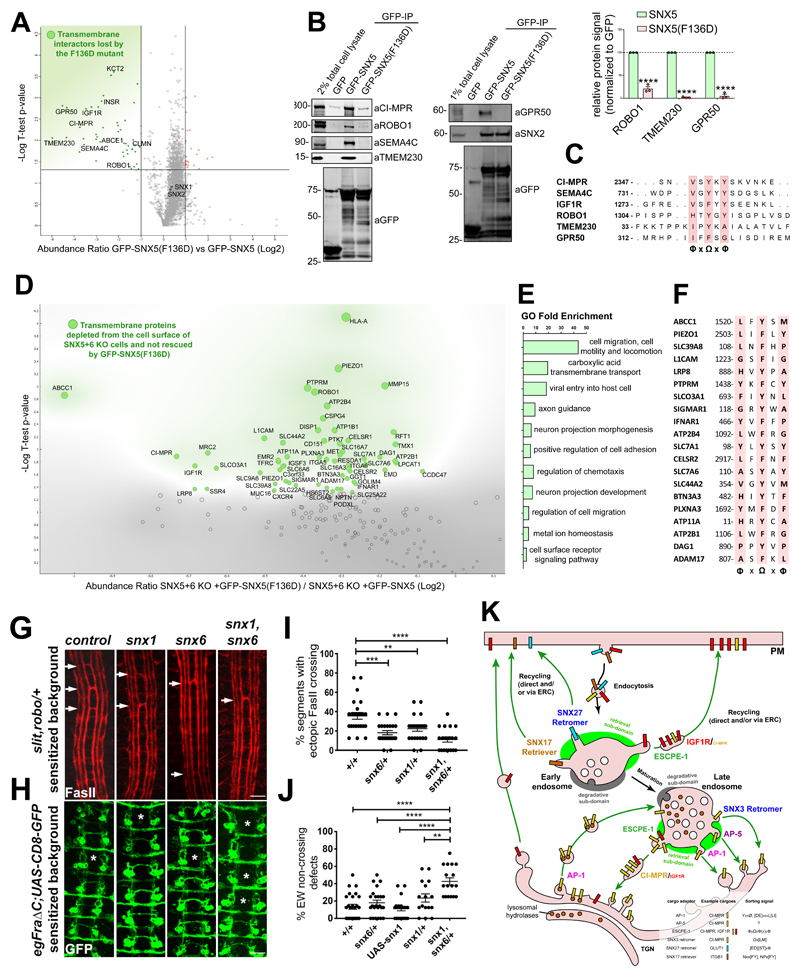
Figure 8. Identification of a ФxΩxФ consensus motif for SNX5-mediated cargo recruitment.
(A) Analysis of comparative interactome of wild-type SNX5 vs SNX5(F136D) mutant across n=3 independent experiments using One-sample t-test and Benjamini-Hochberg FDR. (B) Co-immunoprecipitation of GFP-tagged SNX5 and SNX5(F136D) transiently transfected in HEK293T cells and summary of relative binding. Band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to GFP expression and presented as the relative fraction of the GFP-SNX5 control. Bars, mean; error bars, SEM; circles represent individual data points. Unprocessed scans of immunoblots: Supplementary Figure 3. (C) Alignment of the cytosolic tail of SNX5 cargoes identified through comparative interactome of wild-type SNX5 vs SNX5(F136D) mutant. (D) Comparative proteomic analysis of transmembrane proteins that rely on the ability to engage the SNX5 hydrophobic groove for their plasma-membrane localisation. Analysis was performed across n=3 independent experiments using One-sample t-test and Benjamini-Hochberg FDR. (E) List of most represented GO of the SNX5 cargoes, established by comparative surfaceosome analysis. (F) Alignment of the cytosolic tail of the SNX5 cargoes that require the SNX5 hydrophobic groove for their endosomal retrieval and recycling as established by comparative surfaceosome analysis. (G) Drosophila Snx1 and Snx6 promote axon growth across the midline. Representative images of stage 17 Drosophila embryos stained with anti-fasciclin II (FasII) antibodies to reveal a subset of ipsilaterally projecting interneurons in the CNS. All embryos are heterozygous for mutations in slit and robo. Arrows indicate segments in which axon bundles are abnormally crossing the midline. In embryos that are also heterozygous for either snx1, snx6 or both snx1 and snx6 the percentage of segments showing ectopic midline crossing is reduced compared to control embryos. (H) Representative images of stage 16 embryos stained with anti-GFP to reveal the Eagle subset of commissural interneurons, comprised of Eg axons, which cross the midline in the anterior commissure and Eg axons, which cross in the posterior commissure. Asterisks (*) indicate segments where the EW axons have failed to cross the midline. All embryos selectively express the Fra∆C transgene, which results in the failure of a portion of EW axons to cross the midline. In embryos heterozygous for mutations in both snx1 and snx6 there is a significant reduction in the percentage of EW axons that cross the midline. (I) Quantification of ectopic midline crossing in the indicated genotypes; n=number embryos; wild-type control (n=26), snx6/+ (n=26), snx1/+ (n=24), snx1 (n=23), snx6/+ (n=23). 11 segments were scored in each embryo. Statistical significance was determined using one-way ANOVA (followed by Tukey’s test), **P<0.01, ***P<0.001, ****P<0.0001. (J) Quantification of failed midline crossing in the indicated genotypes; n=number embryos; wild-type control (n=26), snx6/+ (n=24), snx1/+ (n=16), snx1 (n=17), snx6/+ (n=17) and UASSnx1 (n=22). The eight abdominal segments were scored in each embryo. Statistical significance was determined using one-way ANOVA (followed by Tukey’s test), **P<0.01, ***P<0.001, ****P<0.0001. In (I) and (J) each embryo (in which segments were quantified) was considered an independent trial. Scale bar in (G) and (H) 10 microns. (K) A model of the role of the ESCPE-1 complex, which consists of heterodimeric combinations of either SNX5 or SNX6 dimerised to either SNX1 or SNX2, in retrieving and recycling transmembrane cargo protein on the cytosolic facing surface of endosomes. The cartoon also represents the other machineries involved in the retrograde transport of cargoes to the TGN or in their recycling to the plasma membrane. Known sorting motifs within the cytosolic domain of the prototypical cargoes are listed in the figure legend. (H, J) Bars, mean; error bars, SEM; circles represent individual data points. Statistics source data: Supplementary Table 4. Proteomic datasets: Supplementary Table 3.