Abstract
Two new species of Lasiodiplodia (Lasiodiplodia endophytica and Lasiodiplodia magnoliae) are described and illustrated from Magnolia forests in Yunnan, China. Endophytic and saprobic Lasiodiplodia pseudotheobromae and endophytic L. thailandica are new records from this host. The internal transcribed spacers (ITS), part of the translation elongation factor-1α (tef1) and partial β-tubulin (tub2) sequence data were analyzed to investigate the phylogenetic relationships of the new species with other Lasiodiplodia species. Lasiodiplodia magnoliae is phylogenetically sister to L. mahajangana and L. pandanicola but morphologically distinct from L. mahajangana in having larger conidia. Lasiodiplodia endophytica is most closely related to L. iraniensis and L. thailandica and the three species can be distinguished from one another by 2 base pair differences in ITS and three or four base pair differences in tef1. The new collections suggest that Magnolia forest plants are good hosts for Lasiodiplodia species with endophytic and saprobic life-styles.
Subject terms: Fungal biology, Fungal ecology
Introduction
Magnolia species are widely distributed in temperate and tropical South East and East Asia. The wood is used extensively for the interior finish of houses and for door panels, e.g. Magnolia champaca, while the bark of Magnolia officinalis and other species is used in China as a valuable drug1. Many species of Magnolia and their hybrids are cultivated in gardens, grown as temple trees, and the flowers are used for decoration1.
During an investigation of Ascomycetes in sub-tropical regions of Yunnan, China we collected samples from Magnolia trees. Several isolates were of saprobic asexual fungi with hyaline and brown conidia bearing longitudinal striations and conspicuous conidiomatal paraphyses. These initial morphological observations suggested that the isolates are Lasiodiplodia species. Lasiodiplodia Ellis & Everh. is a genus in the family Botryosphaeriaceae (Botryosphaeriales, Dothideomycetes, Ascomycota)2–4 and typified by L. theobromae (Pat.) Griffon & Maubl.3,5–7. Botryosphaeriaceae forms a monophyletic lineage with 22 genera that are defined according to morphology of ascospores and conidia, and phylogenetic relationships4. The family is characterized by large, ovoid to oblong, usually hyaline, aseptate ascospores and hyaline or pigmented, aseptate, one or rarely multi-septate, thick walled conidia usually with longitudinal striations4,7. Considering asexual characters i.e. especially conidia characters, Lasiodiplodia species differ from other closely related genera in the Botryosphaeriaceae by the presence of pycnidial paraphyses and longitudinal striations on mature conidia3 while morphology (especially dimensions) of conidia and paraphyses is used for species delimitation7,8. Some genera of Botryosphaeriaceae show similar morphological affinities to Lasiodiplodia and some morphological characters can be used to distinguish these taxa from Lasiodiplodia7. As an example, Barriopsis species have ovoid conidia with striations even clearly visible in hyaline immature stage as well as pigmented mature stage7,9. The striated, pigmented, mature, ovoid conidia suggest close resemblances to Lasiodiplodia but the early development of striations in hyaline immature stage is a unique character for Barriopsis7,9. Another example is the genus Neodeightonia that shows close affinity to Lasiodiplodia in having striations pigmented mature conidia and can be differentiate from Lasiodiplodia by the absence of conidiomatal paraphyses7,9. However, on account of morphological variability within species, morphology alone is not reliable for distinguishing different Lasiodiplodia species. Phillips et al.4 suggest that combined LSU and ITS provide reliable resolution for phylogeny of Botryosphaeriales. However, protein coding genes such as tef1 and tub2 in addition to LSU and ITS provide greater support for species and genera level delimitation in order Botryosphaeriales4. In previous studies, phylogenetic analyses were solely based on ITS nucleotide sequences3 to identify Lasiodiplodia species. Inclusion of tef1 sequences gives better resolution of phylogenetic relationships among species3,6. The recent multi locus phylogenetic approaches with ITS, tef1 and tub2 nucleotide sequence data has advanced the recognition of numerous Lasiodiplodia species with high phylogenetic support3,4,10,11. In that respect, sequence data of the internal transcribed spacers (ITS), partial translation elongation factor-1α (tef1) and partial β-tubulin (tub2) are now relied on for resolution of species in Lasiodiplodia11. There are 55 epithets of Lasiodiplodia recorded in Index Fungorum (March 2019) and 43 species names in MycoBank (March 2019). However, cultures and DNA sequence data are available for only 35 species (March 2019)10–12.
Species of Lasiodiplodia are cosmopolitan in tropical and subtropical regions and occur on a wide range of monocotyledonous, dicotyledonous and gymnosperm hosts2,3,6,8,13. They exhibit diverse life-styles as endophytes, inhabiting different asymptomatic plant tissues8,14,15, pathogens that cause diseases in various plant hosts3,16 and saprobes that are commonly found on dead woody plant tissues3,17.
To the best of our knowledge there have been no studies on the Lasiodiplodia species associated with Magnolia species in Yunnan Province, China. The aim of this study was to characterize Lasiodiplodia isolates in terms of morphology and phylogeny based on ITS, tef1 and tub2 sequence data.
Results
Phylogenetic analyses
The combined dataset of ITS, tef1 and tub2 consisted of 54 taxa of Lasiodiplodia, with Diplodia mutila (CMW 7060) as the out group taxon and comprised 1267 characters including gaps after alignment. Of these, 1011 were conserved and 123 variable characters were parsimony uninformative. Maximum parsimony analysis of the remaining 133 parsimony informative characters resulted in 1000 equally parsimonious trees of 535 steps with CI = 0.632, RI = 0.798, RC = 0.504 and HI = 0.368. The maximum likelihood analysis resulted in a tree with largely the same topology as the maximum parsimony tree. The RAxML analysis yielded a best scoring tree with the final ML optimization likelihood value of - 4851.693940 (ln). Estimated base frequencies were as follows; A = 0.209292, C = 0.303982, G = 0.256083, T = 0.230643; substitution rates AC = 1.189236, AG = 3.165454, AT = 1.301265, CG = 1.047358, CT = 4.430504, GT = 1.000000; gamma distribution shape parameter α = 0.612671 (Fig. 1).
Figure 1.
Maximum likelihood tree resulting from analysis of the combined ITS, tef1 and tub2 sequence data alignment. Bootstrap values for maximum likelihood (ML, first set) greater than 50, and maximum parsimony (MP, second set) greater than 50 are indicated at the nodes. The tree is rooted with Diplodia mutila (CMW 7060). The newly isolated strains are indicated in red bold and ex-type strains are indicated in black bold.
Four of the isolates from Magnolia clustered with known species, three with Lasiodiplodia pseudotheobromae and one with L. thailandica. The remaining two isolates formed distinct lineages representing two new species. Lasiodiplodia magnoliae was clustered separately and sister to L. mahajangala and L. pandanicola and L. endophytica formed a separate lineage and sister to L. iraniensis and L. thailandica.
Taxonomy
Description of Lasiodiplodia magnoliae N.I. de Silva, A.J.L. Phillips & K.D. Hyde, sp. nov.
Index Fungorum number: IF556217, Faces of Fungi number: FoF 05797 Fig. 2
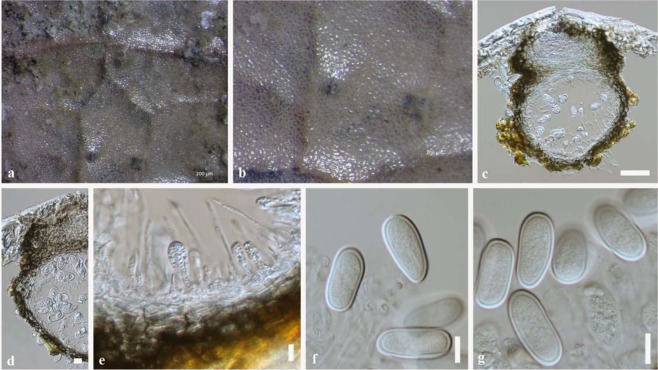
Figure 2.
Lasiodiplodia magnoliae (MFLU 18-1030, holotype). (a,b) Appearance of conidiomata on dead leaf of Magnolia candolii. (c) Vertical section through conidioma. (d) Peridium. (e) Conidiogenous cells and paraphyses. (f,g) Conidia. Scale bars: c = 50 μm, d = 10 μm, e = 5 μm f, g = 10 μm.
Etymology – the epithet “magnoliae” refers to the host plant from which the taxon was collected.
Holotype: MFLU 18-1030
Saprobic on dead leaves attached to the tree of Magnolia candolii Asexual morph: Conidiomata 180–200 µm diam., 200–250 µm high, globose to subglobose, dark brown to black, scattered, solitary, immersed and uniloculate without a conspicuous ostiole. Paraphyses up to 60–70 μm long, 2–4 μm wide, hyaline, cylindrical, septate and rounded at apex. Conidiophores absent. Conidiogenous cells 2–5 µm diam., hyaline, discrete, smooth and cylindrical. Conidia (24–)25–27(–30) × 11–15 μm, hyaline, aseptate, ellipsoid to ovoid, with granular content, both ends broadly rounded, wall <2 μm thick.
Colonies on PDA reaching 30 mm diameter after 1 week at 20–25 °C, colonies medium sparse, circular, flat, surface slightly rough with edge entire, margin well-defined, cottony to fairly fluffy with sparse aspects, colony from above grayish-green to black with fluffy appearance reverse black.
Material examined
China, Yunnan Province, Xishuangbanna, dead leaves attached to the tree of Magnolia candolii (Magnoliaceae), 26 April 2017, N. I. de Silva, NI171 (Holotype MFLU 18-1030; Isotype HKAS100663), ex-type living cultures MFLUCC 18-0948, KUMCC 17-0198.
Notes
The combined ITS, tef1 and tub2 phylogeny showed that Lasiodiplodia magnoliae (MFLUCC 18-0948) clades sister to L. mahajangana and L. pandanicola with low support (53% ML, 60% MP) (Fig. 1). DNA sequence comparisons of ITS and tef1 among L. magnoliae, L. mahajangana and L. pandanicola are given in Table 1. Comparison of total length of 445 bases of ITS sequences revealed one base pair difference among L. magnoliae, L. mahajangana and L. pandanicola. Comparison of total length of 450 bases of tef1 sequences revealed an insertion of eight bases in Lasiodiplodia magnoliae when compared to L. mahajangana and L. pandanicola (Table 1). Lasiodiplodia magnoliae has longer conidia (24–30 μm) than L. mahajangana (14–24 μm)18. Lasiodiplodia pandanicola has overlapping range of conidial dimensions (14–38 μm)10 with L. magnoliae. Lasiodiplodia magnoliae has longer paraphyses (60–70 μm) than L. mahajangana (27–66 μm)17. These three species were found on different host species and with different geographic distributions. Lasiodiplodia magnoliae was isolated from Magnolia candolii in Yunnan, China. Lasiodiplodia mahajangana was isolated from Terminalia catappa in Madagascar18. Lasiodiplodia pandanicola was isolated from dead leaves of Pandanus in Thailand10.
Table 1.
Nucleotide differences between Lasiodiplodia magnoliae (MFLUCC 18-0948) and ex-type isolates of L. mahajangana and L. pandanicola.
| Locus | ITS | tef1- α | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Nucleotide position | 12 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 |
| L. magnoliae MFLUCC 18-0948 (NI171) | G | C | G | G | C | G | C | T | G |
| L. mahajangana | A | — | — | — | — | — | — | — | — |
| L. pandanicola | A | — | — | — | — | — | — | — | — |
Lasiodiplodia pseudotheobromae Alves & Crous, Fungal Diversity 28: 8 (2007). Figs 3 and 4
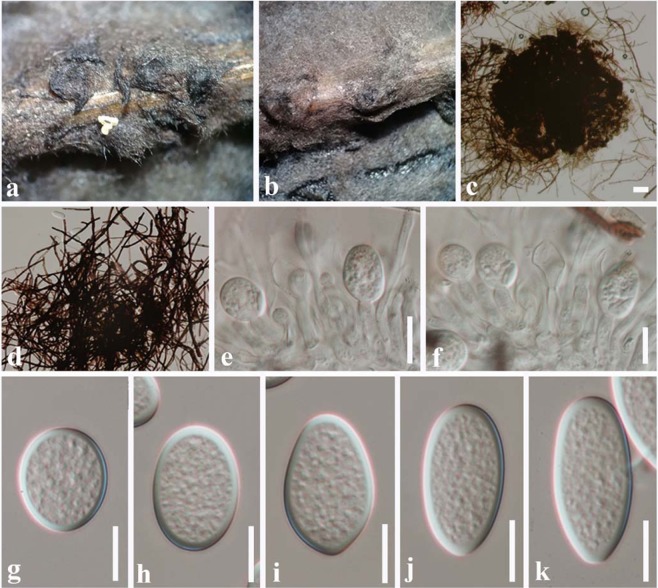
Figure 3.
Lasiodiplodia pseudotheobromae (MFLUCC 18-1120, MFLUCC 18-0950). (a,b) Appearance of conidiomata on twig of Magnolia species. (c,d) Vertical sections through conidiomata. (e) Peridium. (f) Paraphyses (g) Conidiogenous cells. (h–j) Hyaline conidia. (k) Brown conidia on the surface of host. (l,m) Brown conidia. Scale bars: c, d = 50 μm, e, f = 20 μm, g = 5 μm, h–m = 10 μm.
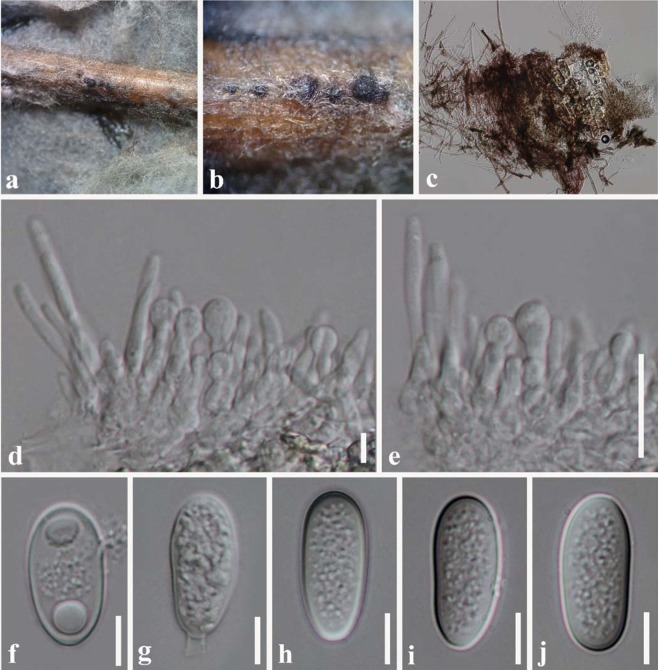
Figure 4.
Lasiodiplodia pseudotheobromae (MFLUCC 18-0951). (a,b) Conidiomata on bamboo sticks in PDA culture plate. (c) Squash mount of conidiomata (d) Mycelium (e,f) Conideogeous cell (g–k) Conidia. Scale bars: c = 50 μm, e, f = 10 μm, g–k = 10 μm.
See Alves et al. (2007).
Material examined
China, Yunnan Province, Xishuangbanna, dead twigs (attached to the tree) of Magnolia candolii (Magnoliaceae), 26 April 2017, N. I. de Silva, NI173 (MFLU 18-1032, HKAS100665), living culture, MFLUCC 18-1120, KUMCC 17-0200; NI173A (HKAS100666), living culture MFLUCC 18-0950, KUMCC 17-0201; Ibid., fresh leaves of Magnolia candolii (Magnoliaceae), 26 April 2017, N. I. de Silva, C13; living culture, MFLUCC 18-0951, KUMCC 17-0218.
Notes
According to the combined ITS, tef1 and tub2 phylogeny, two isolates NI173 and NI173A from M. candolii twigs clustered with Lasiodiplodia pseudotheobromae with low support (51% ML, 52% MP) (Fig. 1). Conidia of these two isolates were hyaline, (20–26 × 10–14 μm) and brown (19–25 × 12–15 μm) and thus are smaller than in the ex-type isolate (27.5–28.5 × 15.5–16.5 μm)6. One endophytic strain (C13) from the same M. candolii plant was phylogenetically closely related to L. pseudotheobromae and clustered with two saprobic strains. Conidial dimensions of the endophytic isolate (26–31 × 10–12 μm) overlap with those of the ex-type isolate. The type of L. pseudotheobromae was isolated from Gmelina arborea in Costa Rica and has also been recorded from Citrus sp., Coffea sp.3, Pteridium aquilinum19, and Plukenetia volubilis20. Here we record endophytic and saprobic L. pseudotheobromae for the first time on Magnolia candolii in Yunnan, China.
Lasiodiplodia thailandica T. Trakunyingcharoen, L. Lombard & Crous, Persoonia 34: 95 (2015) Fig. 5
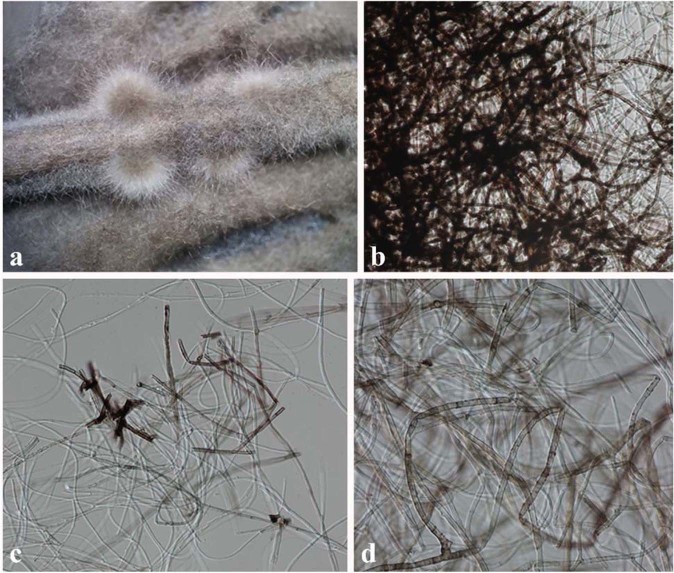
Figure 5.
Lasiodiplodia thailandica (MFLUCC 18-0952). (a,b) Conidiomata on bamboo sticks in PDA culture plate. (c) Squash mount of conidiomata (d) Conideogeous cell (e) Paraphyses (f–j) Conidia. Scale bars: d = 5 μm, e = 20 μm, f–j = 10 μm.
See Trakunyingcharoen et al. (2015).
Material examined
China, Yunnan Province, Xishuangbanna, fresh leaves of Magnolia candolii (Magnoliaceae), 26 April 2017, N. I. de Silva, C17; living culture, MFLUCC 18-0952, KUMCC 17-0222.
Notes
Phylogenetically, the new isolate clustered with the ex-type isolate of Lasiodiplodia thailandica (CBS 138760) based on combined ITS, tef1 and tub2 sequence data. However, the new isolate has larger conidia (28–29 × 11–13 μm) than the ex-type of L. thailandica (20–26 × 12–16 μm)21. Lasiodiplodia thailandica was first described from symptomless twigs of Mangifera indica in Chiang Mai province, Thailand21 and also has been recorded from a petiole of Phyllanthus acidus in Thailand20, from cankered branch of Podocarpus macrophyllus in China19 and from cankered branch of Albizia chinensis in China19.
Lasiodiplodia endophytica N.I. de Silva, A.J.L. Phillips & K.D. Hyde, sp. nov.
Index Fungorum number: IF556218, Faces of Fungi number: FoF 05798 Fig. 6
Figure 6.
Lasiodiplodia endophytica (MFLUCC 18-1121, holotype). (a) Conidiomata on bamboo sticks in PDA culture plate. (b) Squash mount of conidiomata (c,d) Fungal mycelia.
Etymology – the epithet “endophytica” refers to the endophytic life style of this fungus.
Holotype: MFLU 19-0441
Endophytic on fresh leaves of Magnolia candolii. Conidiomata not observed on bamboo sticks on PDA, MEA or Water Agar. Lasiodiplodia endophytica (S8) clusters with L. iraniensis and L. thailandica in a moderately supported clade. It differs from L. iraniensis by unique fixed alleles in two loci: ITS position 463 (C); tef1 positions 554 (C), 599 (T), 681 (C), 703 (G) and differs from L. thailandica: ITS position 463 (C); tef1 positions 551 (C), 598 (C), 671 (C) 811 (C).
Colonies on PDA reaching 30 mm diameter after 3 days at 20–25 °C, colonies medium sparse, circular, surface slightly rough with edge entire, margin well-defined, cottony to fairly fluffy with sparse aspects, colony from above: grey to black with fluffy appearance; reverse black.
Material examined
China, Yunnan Province, Xishuangbanna, fresh leaves of Magnolia candolii (Magnoliaceae), 26 April 2017, N. I. de Silva, S8 (Holotype - a dry culture on bamboo sticks - MFLU 19-0441), living cultures, MFLUCC 18-1121, KUMCC 17-0233.
Notes
The combined ITS, tef1 and tub2 phylogeny showed that Lasiodiplodia endophytica (S8) (MFLUCC 18-1121) clusters sister to Lasiodiplodia iraniensis. DNA sequence comparisons of ITS and tef1 among L. endophytica, L. iraniensis and L. thailandica are given in Table 2. Comparison of total length of 477 bases of ITS sequences revealed one base pair difference among three strains and one base deletion in L. endophytica. Comparison of total length of 290 bases of tef1 sequences revealed seven base pair differences among three strains as given in Table 2. This isolate did not sporulate in culture and no conidiomata were seen on the host. Therefore it was not possible to observe conidial characters. The type of L. iraniensis was isolated from twigs of Salvadora persica in Iran3. Additionally, L. iraniensis was recorded from twigs of Juglans sp. Citrus sp. and Mangifera indica in Iran3.
Table 2.
Nucleotide differences between Lasiodiplodia endophytica (MFLUCC 18-1121) and ex-type isolates of L. iraniensis and L. thailandica.
| Locus | ITS | tef1- α | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Nucleotide position | 420 | 466 | 22 | 25 | 69 | 137 | 138 | 160 | 168 |
| Lasiodiplodia endophytica MFLUCC 18-1121 (S8) | C | — | C | C | T | C | C | G | C |
| Lasiodiplodia iraniensis | T | T | C | A | A | C | T | C | C |
| Lasiodiplodia thailandica | T | T | A | C | T | T | C | G | T |
Discussion
In this study two new species of Lasiodiplodia were identified and described from Magnolia candolii in the southern part of Yunnan Province, China. One species (Lasiodiplodia magnoliae) was considered to be saprobic, while the other (L. endophytica) was thought to be endophytic. In addition, two saprobic isolates of L. pseudotheobromae from dead twigs and an endophytic isolate of the same species from fresh leaves of Magnolia candolii were recorded for the first time from China. An endophytic isolate of Lasiodiplodia thailandica was also isolated for the first time from fresh leaves of Magnolia candolii in China. When Promputtha et al.22 studied endophytes and Promputtha et al.23 studied saprobes from leaf litter of Magnolia liliifera and M. garretii respectively in Chiang Mai, Thailand, no Lasiodiplodia species were recorded.
Phylogenetic approaches based on DNA sequence data have played a significant role in distinguishing species in Lasiodiplodia3,4,24,25. Previous studies have used combined ITS and tef1 regions to clarify the taxonomy and phylogenetic relationships of species in Lasiodiplodia3,6,26 while others have used combined ITS, tef1, tub2 and rpb219. The current phylogenetic analyses with combined ITS, tef1 and tub2 sequence data gave good resolution of phylogenetic separations among Lasiodiplodia species and provide insights in to taxonomic novelties. Unfortunately, amplification of tef1 of MFLUCC 18-0951 - C13 and MFLUCC 18-0952 - C17, and tub2 of MFLUCC 18-0951 - C13 was not successful in this study. Phylogenetic analyses were conducted using DNA sequence data available in GenBank, but unfortunately sequences of tef1 and tub2 are not available for some species (see Table S1) and some of the sequences are shorter than expected. These issues of tef1 and tub2 might compromise the number of characters in the final alignment and ultimately might affect the final phylogenetic tree construction. We provide phylogenetic analyses for single molecular markers as Supplementary Materials. Phylogenetic trees from ITS and tub2 did not provided good resolution among Lasiodiplodia species. The phylogenetic analysis of ITS gene showed that three newly isolated strains of Lasiodiplodia pseudotheobromae and L. thailandica MFLUCC 18-095 clustered in one group and L. endophytica MFLUCC 18-1121 clustered separately from that group. Lasiodiplodia magnoliae MFLUCC 18-0948 formed a separate clade with L. citricola IRAN1522C. The phylogenetic analysis of tub2 did not provide clear separation of newly isolated strains and previously described species. Two Lasiodiplodia pseudotheobromae strains, L. thailandica and L. endophytica clustered together and L. magnoliae formed a separate, distantly related lineage. Analysis of tef1 resulted in a better resolution of many taxa than single ITS and tub2 gene trees and showed similar phylogenetic relationships as combined ITS, tef1 and tub2 analyses. In both tef1 gene and combined ITS, tef1 and tub2 gene phylogenetic analyses, new strains of Lasiodiplodia pseudotheobromae formed a clade with other L. pseudotheobromae, L. endophytica clustered with L. iraniensis, L. thailandica and L. hyalina. In both tef1 gene and combined phylogenetic analyses, Lasiodiplodia magnoliae reflected similar phylogenetic affiliation with L. chonburiensis, L. caatinguensis, L. exigua, L. pandanicola and L. mahajangana. It is also worth noting that the phylogenetic relationships of species within Lasiodiplodia recovered herein from combined ITS, tef1 and tub2 gene analyses are similar to previously established ones in Dissanayake et al.12, Dou et al.11 and Tibpromma et al.10. These three phylogenetic studies were based on different combinations of molecular markers such as Dissanayake et al.12 who used combined ITS and tef1, Dou et al.11 used combined ITS, tef1, tub2 and rpb2 and Tibpromma et al.10 used ITS, tef1 and tub2. In earlier studies, Lasiodiplodia were species distinguished solely on their ITS sequences10. In recent studies, taxonomists frequently use highly variable protein coding genes such as tef1, tub2 together with ITS to construct phylogenies especially at species levels4. It can be assumed that these combination of molecular markers strengthen the support for them and to separate the existing ones3,4.
Conidia of Lasiodiplodia species are initially hyaline, aseptate, ellipsoid to ovoid and become pigmented, 1-septate with longitudinal striations3. We observed hyaline, aseptate conidia and brown, 1-septate conidia with longitudinal striations in the saprobic isolates of Lasiodiplodia pseudotheobromae, but only hyaline conidia were seen in Lasiodiplodia magnoliae, the endophytic isolate of L. pseudotheobromae and L. thailandica. Previous studies have recorded both hyaline and pigmented conidia in L. pseudotheobromae6 and L. thailandica21. Other Lasiodiplodia species have been observed with only hyaline conidia such as L. chonburiensis10, L. sterculiae27 and L. thailandica in which most conidia were hyaline and only 10% were brown19. This character does not seem to be restricted to any particular phylogenetic groups but appears in different Lasiodiplodia species. Thus, L. magnoliae and L. chonburiensis are closely related and found in one clade. On the other hand, L. sterculiae and L. thailandica are distantly related to both L. magnoliae and L. chonburiensis and formed widely separate lineages in the phylogenetic tree. Lasiodiplodia magnoliae differs from its sister taxa by phylogeny, morphology, host species and locality as described in the notes section that support for the introduction of new saprobic taxa. We were unable to observe conidia of L. endophytica (S8) in culture even after many attempts on different media. It was considered here that phylogeny based on combined ITS, tef1 and tub2 sequence data provides sufficient evidence for the designation of L. endophytica (S8) as a novel taxon.
Lasiodiplodia species exhibit diverse life-styles as endophytes8,15, pathogens3,16 and saprobes3,17. Lasiodiplodia species with endophytic life-styles are associated with different asymptomatic plant tissues such as L. avicenniae from asymptomatic branches of Avicennia marina in South Africa, L. bruguierae from asymptomatic branches of Bruguiera gymnorrhiza in South Africa28 and L. mahajangana from healthy branches of Terminalia catappa in Madagascar18. We isolated three endophytic species; Lasiodiplodia endophytica, L. pseudotheobromae and L. thailandica from asymptomatic leaves of Magnolia candolii. Interestingly, we isolated one endophytic and 2 saprobic isolates of Lasiodiplodia pseudotheobromae. This might be possible because endophytes switch their nutritional mode to saprobic when environmental conditions become unfavorable to the host or during host senescence29. Thus, de Errasti et al.30 stated that diatrypaceous endophytic fungi switch to a saprotrophic life-style during host senescence. They explained that this might be ecologically important as they can decay the plant part when it dies30. In another scenario, Osorio et al.28 showed that endophytic Lasiodiplodia avicenniae became pathogenic and caused lesions on the branches of Avicennia marina after inoculating. It is assumed that some fungi exhibit a continuum of life-styles ranging from biotrophy (or endophytic), through to necrotrophy and ultimately to saprotrophy29. Endophytes are a hidden bioresource of fungal diversity that have the potential to produce important bioactive agents15. Chen et al.15 chemically investigated a strain of Lasiodiplodia sp. isolated from asymptomatic leaves of the medicinal plant Acanthus ilicifolius. They studied β-resorcylic acid derivatives and showed that these compounds showed more potent inhibitory effects against α-glucosidase activity than the clinical α-glucosidase inhibitor acarbose15. Lasiodiplodia species with pathogenic life-styles are associated with shoot blights, stem cankers, fruit rots, dieback, grapevine trunk diseases and gummosis3,16,31 Lasiodiplodia exigua from a branch canker of Retama raetam32, L. mediterranea from branch canker of Quercus ilex32, L. plurivora from V-shaped necrotic lesion of Prunus salicina, in Africa33 and L. pseudotheobromae from grapevine trunk disease16 are some examples that cause different plant diseases. We did not observe any pathogenic Lasiodiplodia species in our study. Saprobic Lasiodiplodia species have been recorded such as Lasiodiplodia iraniensis on dead twigs of Salvadora persica, L. hormozganensis on Olea sp.3 and L. theobromae on dead twigs of Eucalyptus sp.17. Similarly, we introduced a new saprobic species of Lasiodiplodia magnoliae and two isolates of L. pseudotheobromae.
This study identified Lasiodiplodia species in forest plants of Magnolia candolii in Yunnan, China. The study has expanded the knowledge of Lasiodiplodia species providing two novel species and two new host records. It might be possible to identify new distribution and host associations of Lasiodiplodia species from other forest plants in the world. It is important to study endophytic Lasiodiplodia species as well as pathogenic and saprobic life-styles as novel endophytes are also yet to be explored.
Materials and Methods
The study area was a sub-tropical rain forest inside the Xishuangbanna tropical botanical garden in Xishuangbanna at 21°55′N, 101°15′E, Yunnan province, China. Elevation ranges from 709–869 m and mean temperature and precipitation are 21.0 °C and 1532 mm respectively. The wet season is from May to October while the dry season is from November to April34,35.
Isolation of fungal endophytes
Fresh leaves of Magnolia candolii were collected randomly on 26th of April 2017. The leaves were kept at 4 °C in sterile polyethylene bags until they were processed in the laboratory. Isolation of endophytes was done according to the methods described by Promputtha et al.36 with modifications. First leaves were washed using tap water and cut in to small pieces of leaves (5 × 5 mm2) and soaked in distilled water for 1 minute and then surfaced sterilized by dipping in 70% alcohol followed by 2% NaOCl for 30 s and finally washed with sterile distilled water for 30 s, dried and plated on Potato Dextrose Agar (PDA). Fungi were isolated into pure culture and grouped according to their culture morphology.
Isolation of fungal saprobes
Fungi were isolated from dead twigs attached to the host. Specimens were taken to the laboratory in Ziplock plastic bags and observed with a JNOEC JSZ4 stereomicroscope. Micro-morphological characters were examined with an OLYMPUS SZ61 compound microscope and images recorded with a Canon EOS 600D digital camera mounted on a Nikon ECLIPSE 80i compound microscope. All microscopic measurements were made with Tarosoft (R) image framework v. 0.9.0.7 and images for publication were processed with Adobe Photoshop CS3 extended version. Pure cultures of the fungus were prepared by single spore isolation37. Germinating conidia were transferred aseptically to potato dextrose agar (PDA). Growth rate and colony characteristics were determined from cultures grown on PDA at room temperature (25 °C) for one week.
The specimens cited in this paper are maintained at the Mae Fah Luang University Herbarium (MFLU), Chiang Rai, Thailand and Kunming Institute of Botany herbarium (HKAS), Kunming, China. Cultures were deposited at Kunming Institute of Botany Culture Collection (KUMCC). Faces of Fungi numbers and Index Fungorum numbers were registered as described in Jayasiri et al.38 and Index Fungorum (2019)39.
DNA extraction, PCR amplification and phylogenetic analysis
Mycelium was grown on PDA for one week at 25 °C in normal light in the laboratory. Genomic DNA was extracted from the mycelium using a Biospin fungus genomic DNA kit (BioFlux®, P.R. China) following the manufacturer’s protocol. DNA was kept at 4 °C for DNA amplification and maintained at −20 °C for long term storage.
The internal transcribed spacer (ITS) was amplified with primer pair ITS4 and ITS540 as described in Alves et al.41. Part of the translation elongation factor (tef1) was amplified with primer pair EF1-728F and EF1-986 Carbone and Kohn42 and EF1-688F and EF1-1251R Alves et al.6. The partial β-tubulin (tub2) was amplified with primer pair Bt2a and Bt2b43. The expected sequence lengths are approximately 500 bp, 300–400 bp, 400 bp for ITS, tef1 and tub2 respectively. Quality of PCR products was checked on 1% agarose electrophoresis gels stained with ethidium bromide. The amplified PCR fragments were sequenced by Sangon Biotech (Shanghai) Co., Ltd, P.R. China.
Newly generated nucleotide sequences were deposited in GenBank (Table S1 in Supplementary material). Sequences of the individual loci of ITS, tef1 and tub2 were aligned with MAFFT v. 7 online version44 using default settings. BioEdit v. 7.0.5.245 was used to refine the alignments manually where necessary and to exclude incomplete portions at the ends of the sequences before the analyses.
Maximum likelihood analysis was performed with RAxML GUI v. 1.346 and maximum parsimony analysis was done with PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b1047. Evolutionary models for phylogenetic analyses were selected independently for each locus using MrModeltest v. 3.748 under the Akaike Information Criterion (AIC). GTR + I + G model of nucleotide substitution was selected for the maximum likelihood (ML) analysis. Parameters for maximum likelihood were set to rapid bootstrapping and the analysis carried out using 1000 replicates. Maximum parsimony was run with the heuristic search option, random taxon addition, tree bisection-reconnection (TBR) for the branch swapping algorithm and 1000 random sequence additions, with maxtrees set at 1000. Gaps were treated as missing data. Tree Length [TL], Consistency Index [CI], Retention Index [RI], Relative Consistency Index [RC] and Homoplasy Index [HI] were calculated for the most parsimonious tree. Phylograms were visualized with FigTree v1.4.0 Rambaut49 and annotated in Microsoft Power Point (2010). The alignment and tree files were submitted to TreeBASE with reviewer’s link (http://purl.org/phylo/treebase/phylows/study/TB2: S23955).
Supplementary information
GenBank accession number and single gene phylogenetic trees
Acknowledgements
This work was supported by grants from Chiang Mai University and TRF Research-Team Association Grant (RTA5880006). We wish to thank the Key Research Program of Frontier Sciences, CAS (Grant No. QYZDY-SSWSMC014” and “973 key project of the National Natural Science Foundation of China (grant no. 2014CB954101). The Research Fund from China Postdoctoral Science Foundation (Grant No. Y71B283261), the Yunnan Provincial Department of Human Resources and Social Security (grant no. Y836181261) and the National Nature Science Foundation of China (NSFC; Grant No. 31850410489) is also acknowledged. Germplasm Bank of Wild Species in Southwest China, Kunming Institute of Botany, Chinese Academy of Science, Kunming is thanked for supporting DNA molecular experiments of this study. Dr. Shaun Pennycook is thanked for checking species name. K.D. Hyde acknowledges the Thailand Research Fund for a grant no RDG6130001, entitled Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion and Chiang Mai University for a position as an adjunct Professor. Alan J.L. Phillips acknowledges the support from UID/MULTI/04046/2019 Research Unit grant from FCT, Portugal to BioISI. J.K. Liu would like to thank the National Natural Science Foundation of China (NSFC 31600032) and Science and Technology Foundation of Guizhou Province (LH [2015]7061).
Author Contributions
N.I.D. and K.D.H. designed the study. N.I.D. performed the morphological study and phylogenetic data analyses with help from A.J.L.P. and J.K.L. S.L. provided funding for the study. N.I.D. wrote the manuscript and A.J.L.P., J.K.L., K.D.H. and S.L. reviewed and edited the manuscript. All authors reviewed and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50804-x.
References
- 1.Nooteboom HP, Chalermglin P. The Magnoliaceae of Thailand. Thai Forest Bulletin (Botany) 2009;37:111–138. [Google Scholar]
- 2.Barr, M. E. Prodromus to class Loculoascomycetes. Amherst, Massachusetts: published by the author. (1987).
- 3.Abdollahzadeh J, Javadi A, Goltapeh EM, Zare R, Phillips AJL. Phylogeny and morphology of four new species of Lasiodiplodia from Iran. Persoonia. 2010;25:1–10. doi: 10.3767/003158510X524150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips AJ, Hyde KD, Alves A, Liu JK. Families in Botryosphaeriales: a phylogenetic, morphological and evolutionary perspective. Fungal Divers. 2019;94:1–22. doi: 10.1007/s13225-018-0416-6. [DOI] [Google Scholar]
- 5.Sutton, B. C. The Coelomycetes, Fungi Imperfecti with Pycnidia, Acervuli and Stromata. Commonwealth Mycological Institute, Kew, Surrey, England. (1980).
- 6.Alves A, Crous PW, Correia A, Phillips AJL. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers. 2008;28:1–13. [Google Scholar]
- 7.Phillips AJL, et al. The Botryosphaeriaceae: genera and species known from culture. Studies in Mycology. 2013;76:51–167. doi: 10.3114/sim0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slippers B, Wingfield M. J. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biol Rev. 2007;21:90–106. doi: 10.1016/j.fbr.2007.06.002. [DOI] [Google Scholar]
- 9.Phillips AJL, et al. Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia. 2008;21:29–55. doi: 10.3767/003158508X340742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tibpromma S, et al. Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Divers. 2018;93:1–160. doi: 10.1007/s13225-018-0408-6. [DOI] [Google Scholar]
- 11.Dou, Z. P., He, W. & Zhang, Y. Does morphology matter in taxonomy of Lasiodiplodia? An answer from Lasiodiplodia hyalina sp nov. Mycosphere8, 1014–1027, 10.5943/mycosphere/8/2/5 (2017a).
- 12.Dissanayake, A. J., Phillips, A. J. L., Li, X. H. & Hyde, K. D. Botryosphaeriaceae: Current status of genera and species. Mycosphere7, 1001–1073, 10.5943/mycosphere/si/1b/13 (2016).
- 13.Punithalingam, E. Plant diseases attributed to Botryodiplodia theobromae. Pat. J. Cramer. Vaduz. (1980).
- 14.Johnson GI, Mead AJ, Cooke AW, Dean JR. Mango stem end rot pathogens – Fruit infection by endophytic colonistion of the inflorescence and pedicel. Ann. Appl. Biol. 1992;120:225–234. doi: 10.1111/j.1744-7348.1992.tb03420.x. [DOI] [Google Scholar]
- 15.Chen S, et al. β-Resorcylic acid derivatives with α-glucosidase inhibitory activity from Lasiodiplodia sp. ZJ-HQ1, an endophytic fungus in the medicinal plant Acanthus ilicifolius. Phytochem Lett. 2015;13:141–146. doi: 10.1016/j.phytol.2015.05.019. [DOI] [Google Scholar]
- 16.Dissanayake AJ, et al. Lasiodiplodia pseudotheobromae causes pedicel and peduncle discolouration of grapes in China. Australas. Plant Dis. Notes. 2015;10:21. doi: 10.1007/s13314-015-0170-5. [DOI] [Google Scholar]
- 17.Liu JK, et al. Towards a natural classification of Botryosphaeriales. Fungal Divers. 2012;57:149–210. doi: 10.1007/s13225-012-0207-4. [DOI] [Google Scholar]
- 18.Begoude BD, Slippers B, Wingfield MJ, Roux J. Botryosphaeriaceae associated with Terminalia catappa in Cameroon, South Africa and Madagascar. Mycological Progress. 2010;9:101–123. doi: 10.1007/s11557-009-0622-4. [DOI] [Google Scholar]
- 19.Dou, Z. P., He, W. & Zhang, Y. Lasiodiplodia chinensis, a new holomorphic species from China. Mycosphere8, 521–530 10.5943/mycosphere/8/2/3 (2017b).
- 20.Tennakoon DS, et al. Sexual morph of Lasiodiplodia pseudotheobromae (Botryosphaeriaceae, Botryosphaeriales, Dothideomycetes) from China. Mycosphere. 2016;7:990–1000. doi: 10.5943/mycosphere/si/1b/119. [DOI] [Google Scholar]
- 21.Trakunyingcharoen T, et al. Caulicolous Botryosphaeriales from Thailand. Persoonia. 2015;34:87–99. doi: 10.3767/003158515X685841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Promputtha I, Jeewon R, Lumyong S, McKenzie EH, Hyde KD. Ribosomal DNA fingerprinting in the identification of non-sporulating endophytes from Magnolia liliifera (Magnoliaceae) Fungal Divers. 2005;20:167–186. [Google Scholar]
- 23.Promputtha I, Lumyong S, Lumyong P, McKenzie EC, Hyde KD. Fungal succession on senescent leaves of Manglietia garrettii in Doi Suthep-Pui National park, northern Thailand. Fungal Divers. 2002;10:89–100. [Google Scholar]
- 24.Coutinho IBL, et al. Diversity of genus Lasiodiplodia associated with perennial tropical fruit plants in northeastern Brazil. Plant Pathology. 2017;66:90–104. doi: 10.1111/ppa.12565. [DOI] [Google Scholar]
- 25.Netto MS, et al. Analysis of phylogeny, distribution, and pathogenicity of Botryosphaeriaceae species associated with gummosis of Anacardium in Brazil, with a new species of Lasiodiplodia. Fungal biology. 2017;121:437–451. doi: 10.1016/j.funbio.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Phillips AJL, Alves A, Correia A, Luque J. Two new species of Botryosphaeria with brown, 1-septate ascospores and Dothiorella anamorphs. Mycologia. 2005;97:513–529. doi: 10.1080/15572536.2006.11832826. [DOI] [PubMed] [Google Scholar]
- 27.Yang T, et al. Families, genera and species of Botryosphaeriales. Fungal Biology. 2016;121:322–346. doi: 10.1016/j.funbio.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Osorio JA, Crous CJ, De Beer ZW, Wingfield MJ, Roux J. Endophytic Botryosphaeriaceae, including five new species, associated with mangrove trees in South Africa. Fungal Biology. 2017;121:361–393. doi: 10.1016/j.funbio.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 29.De Silva NI, et al. Mycosphere Essays 9: Defining biotrophs and hemibiotrophs. Mycosphere. 2016;7:545–559. doi: 10.5943/mycosphere/7/5/2. [DOI] [Google Scholar]
- 30.de Errasti A, Novas MV, Carmarán CC. Plant-fungal association in trees: Insights into changes in ecological strategies of Peroneutypa scoparia (Diatrypaceae) Flora-Morphology, Distribution, Functional Ecology of Plants. 2014;209:704–710. doi: 10.1016/j.flora.2014.07.006. [DOI] [Google Scholar]
- 31.Arx, J. A. von. Plant-pathogenic fungi. Cramer, Berlin. (1987).
- 32.Linaldeddu BT, et al. Diversity of Botryosphaeriaceae species associated with grapevine and other woody hosts in Italy, Algeria and Tunisia, with descriptions of Lasiodiplodia exigua and Lasiodiplodia mediterranea sp. nov. Fungal Divers. 2015;71:201–214. doi: 10.1007/s13225-014-0301-x. [DOI] [Google Scholar]
- 33.Damm U, Crous PW, Fourie PH. Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora sp. nov. Mycologia. 2007;99:664–680. doi: 10.1080/15572536.2007.11832531. [DOI] [PubMed] [Google Scholar]
- 34.Cao M, Zou X, Warren M, Zhu H. Tropical forests of Xishuangbanna, China. Biotropica. 2006;38:306–309. doi: 10.1111/j.1744-7429.2006.00146.x. [DOI] [Google Scholar]
- 35.Aluthwattha ST, et al. Does spatial variation in predation pressure modulate selection for aposematism? Ecology and evolution. 2017;7:7560–7572. doi: 10.1002/ece3.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Promputtha I, et al. Phylogenetic evaluation of whether endophytes become saprotrophs at host senescence. Microbial Ecology. 2007;53:579–590. doi: 10.1007/s00248-006-9117-x. [DOI] [PubMed] [Google Scholar]
- 37.Chomnunti P, et al. The Sooty Moulds. Fungal Divers. 2014;66:1–36. doi: 10.1007/s13225-014-0278-5. [DOI] [Google Scholar]
- 38.Jayasiri SC, et al. The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015;74:3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- 39.Index Fungorum, www.indexfungorum.org 2019 (2019).
- 40.White T, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 1990;18:315–322. doi: 10.1016/b978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- 41.Alves A, Correia A, Luque J, Phillips A. Botryosphaeria corticola sp. nov. on Quercus species, with notes and description of Botryosphaeria stevensii and its anamorph, Diplodia mutila. Mycologia. 2004;96:598–613. doi: 10.1080/15572536.2005.11832956. [DOI] [PubMed] [Google Scholar]
- 42.Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 43.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied & Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada KD, Tomii K, Katoh K. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics. 2016;32:3246–3251. doi: 10.1093/bioinformatics/btw412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 46.Silvestro D, Ingo M. raxmlGUI: a graphical front-end for RAxML. Organisms Diversity and Evolution. 2012;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 47.Swofford, D. L. PAUP: phylogenetic analysis using parsimony, version 4.0 b10. Sinauer Associates, Sunderland, 10.1186/1471-2164-3 (2002).
- 48.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 49.Rambaut, A. FigTree version 1.4.0. Available at, http://tree.bio.ed.ac.uk/software/figtree (accessed 1 January 2019) (2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GenBank accession number and single gene phylogenetic trees