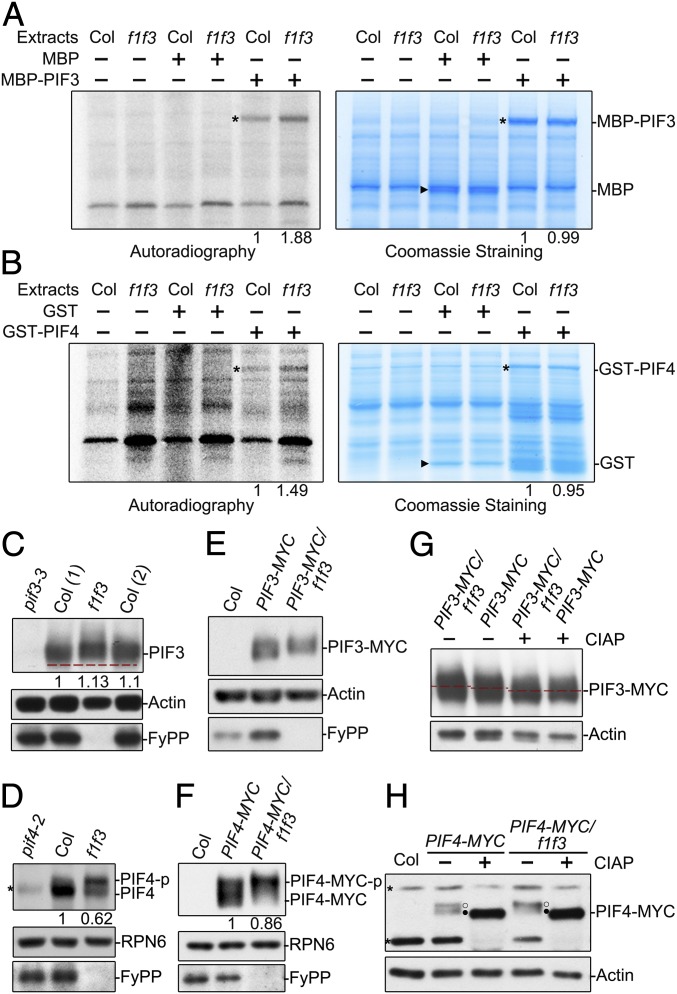
Fig. 3.
PP6 regulated the phosphorylation status of PIF3 and PIF4 in the dark. Cell-free kinase assays showed that there were more phosphorylated MBP-PIF3 (A) and GST-PIF4 (B) proteins in total extracts of dark-grown f1 f3 seedlings than in total extracts of dark-grown Col-0 seedlings. (Left) [γ-32P] adenosine 5′-triphosphate autoradiography. (Right) Coomassie Blue staining. Asterisks denote the bands of GST-PIF4 or MBP-PIF3. Triangles denote the GST or MBP bands. The phosphorylated signals of recombinant PIF proteins incubated with Col were set as 1. Endogenous PIF3 proteins (C) and PIF4 proteins (D) from f1 f3 mutants migrated more slowly than those from Col-0 on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Immunoblotting analyses were performed with anti-PIF3, anti-PIF4, and anti-FyPP antibodies. The numbers in parentheses represents biological replicates. PIF3-MYC proteins (E) and PIF4-MYC proteins (F) from f1 f3 mutants migrated more slowly on SDS-PAGE gels than those from Col-0. Anti-MYC and anti-FyPP antibodies were used in the immunoblotting analyses. In C, D, and F, PIF3, PIF4, and PIF4-MYC protein levels were normalized to RPN6, and PIF protein levels in Col background were set as 1. The slow-migrating PIF3-MYC (G) and PIF4-MYC (H) were phosphorylated forms. Proteins extracted from dark-grown seedlings were incubated with (+) or without (−) alkaline phosphatase (CIAP) for 2 h before electrophoresis. Actin and RPN6 proteins were used as loading controls. Asterisks denote nonspecific bands. Open circles (○) represent the phosphorylated form, and filled circles (●) indicate the dephosphorylated form.