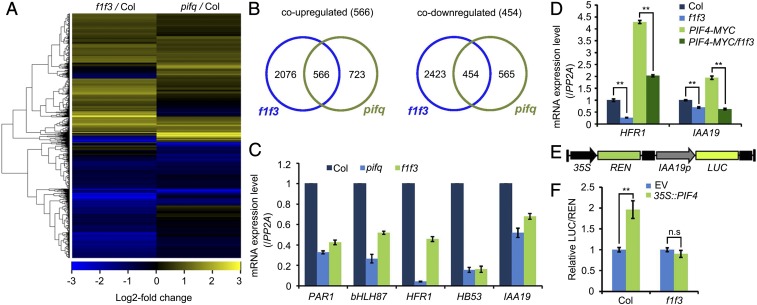
Fig. 5.
PP6 mediated the PIF-regulated transcriptomic changes in the dark-grown seedlings. (A) Heat map of genes up- or down-regulated in f1 f3 and pifq mutants. Up to 44% of PIF-regulated genes were also regulated by PP6. (B) Venn diagrams showing the overlap of genes that were up- or down-regulated in f1 f3 and pifq mutants. (C) RT-qPCR validation of several genes that were coregulated in f1 f3 and pifq mutants. These genes were chosen for validation because they were bound and regulated by PIF4. (D) Relative expression levels of HFR1 and IAA19 in Col-0, f1 f3, PIF4-MYC, and PIF4-MYC/f1 f3 seedlings grown in the dark for 4 d. The expression of PP2A was used as an internal control. Data are mean ± SD (n = 3). **P < 0.01, as calculated by Student’s t test. (E) Structure of the IAA19p::LUC reporter. The positions of the 35S promoter, REN luciferase coding sequence (REN), IAA19 promoter (IAA19p, −2 kilobases to 0 base pair) and firefly luciferase coding sequence (LUC) are indicated. (F) Relative LUC activities (LUC/REN) in Col-0 and f1 f3 mesophyll protoplasts cotransformed with IAA19p::LUC (reporter) and 35S:PIF4 (effector). The empty vector (EV) was used as a reporter control. Data are shown as mean ± SD (n = 4). **P < 0.01, as calculated by Student’s t test. n.s, not significant.