Fig. 3.
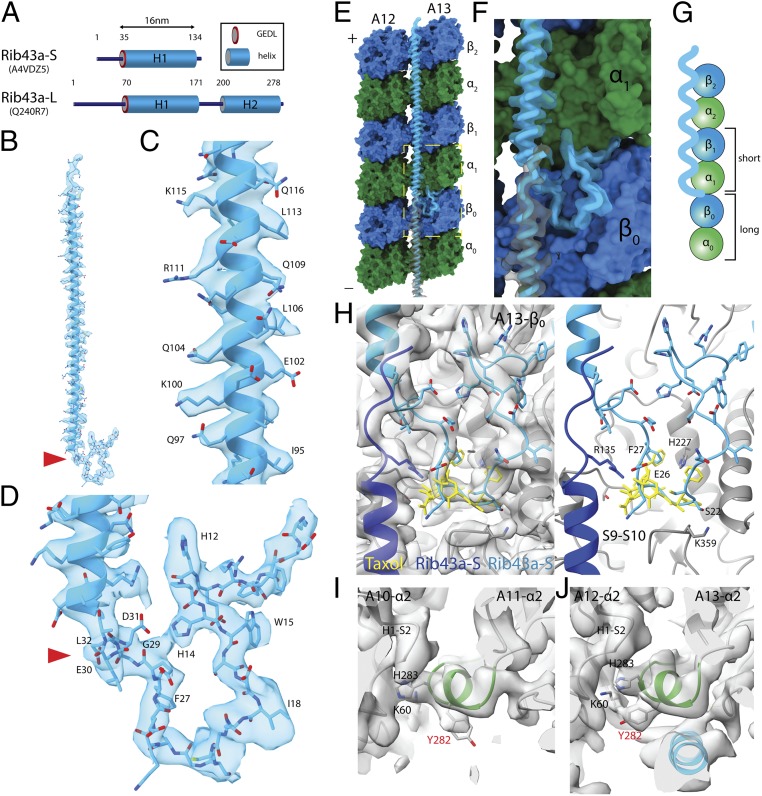
Rib43a leads to the bimodal distance in the PF regions. (A) Secondary structure prediction of Rib43a-S and Rib43a-L. Only the large stretch of α-helices more than 20 residues from the structure prediction is shown. The GEDL consensus sequence is a conserved region of Rib43a (PFAM PF05914). (B) Model of the Rib43a-S inside its segmented density. (C and D) Magnified views of the helical region (C) and the N-terminal region (D) of Rib43a-S. The location of the GEDL motif is shown by the red arrowheads in B and D. (E) Model of Rib43a binds to the PF pair A12/A13. Yellow dashed box shows the magnified view in F. (F) The N terminus of Rib43a-S inserts into the interdimer interface in PF A13. (G) Schematic model of how Rib43a-S binds to the PF leading to the bimodal dimer distance pattern. (H) Superimposed views of taxol (PDB: 5SYF, yellow) and Rib43a-S with map (Left) and without map (Right) show similar topology. R135 in the C terminus of the lower Rib43a-S (dark blue) might interact with E26 of the N terminus of the upper Rib43a-S (light blue) in a head-to-tail dimerization mechanism. (I and J) M-loop conformations in the lateral interaction with Rib43a (PFs A12 and A13) and without (PFs A10 and A11). The side chain of Y282 adopts a different conformation in the presence of Rib43a, potentially due to steric clash. In this conformation, Y282 might interact with K60 of the neighboring α-tubulin.