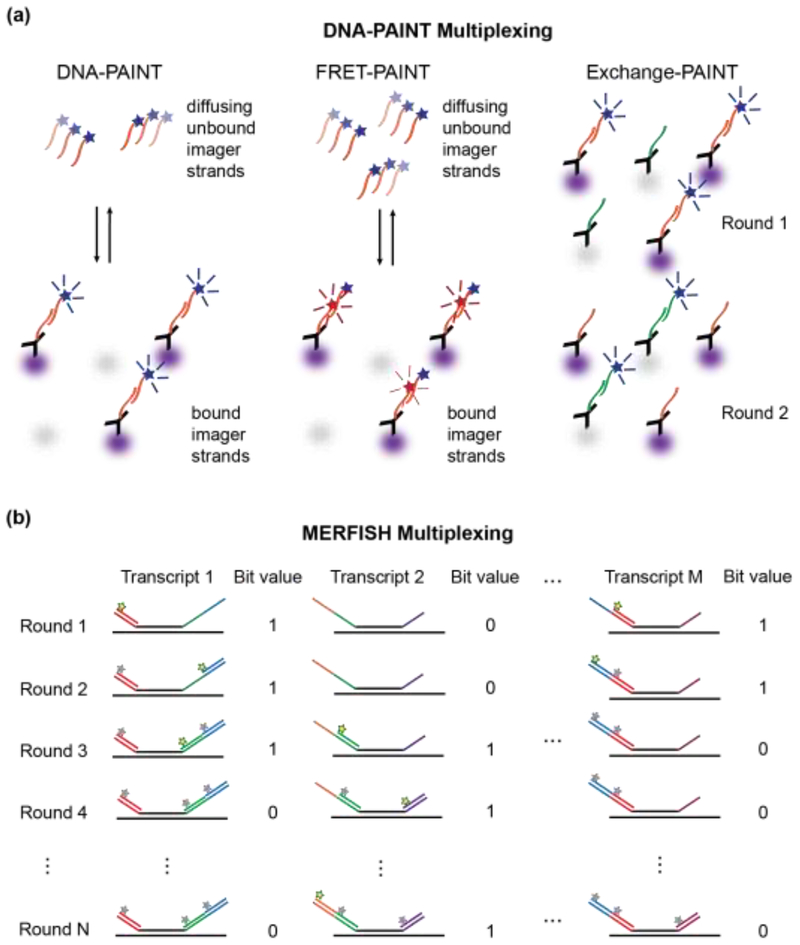
Figure 3. High-content super-resolution microscopy labeling techniques.
a.) DNA-PAINT is used to localize antibody-labeled proteins (purple and gray). Left Panel: Diffusing dye-conjugated imager strands can bind to complementary docking strands on the antibodies. Upon binding, their fluorescence images on the detector become localized in space, which enables precise position estimations of the so-labeled proteins. Middle Panel: Docking strands used in FRET-PAINT are conjugated to a FRET acceptor dye. FRET occurs only after binding of an imager strand, which is conjugated to a FRET donor dye. FRET emission of the acceptor dye is used to localize the labeled protein. Raising the concentration of imager strands does not increase the background in the emission channel of the acceptor dye, but accelerates the data acquisition time by increasing the imager strand binding rates. Right Panel: Exchange-PAINT enables multiplexed labeling by sequentially introducing multiple different imager strands that are conjugated with the same dye after repeated washing steps. b) MERFISH localizes multiple RNA transcripts by introducing encoding RNA probes that bind to M distinct RNA transcripts through complementary base-pairing (black lines). Each encoding probe contains a unique combination of readout sequences (colored lines), which act as docking sites for STORM dye-modified imager probes. Fluorescent imager probes (yellow stars) are sequentially introduced, imaged, and then photobleached or chemically cleaved (grey stars) prior to the next round of imaging. If a localized fluorescence signal is measured in a given imaging round, the position of the RNA transcript can be determined and the bit value for this position is set to 1. Over N rounds of imaging, an N-bit barcode is generated that uniquely identifies the RNA transcript.