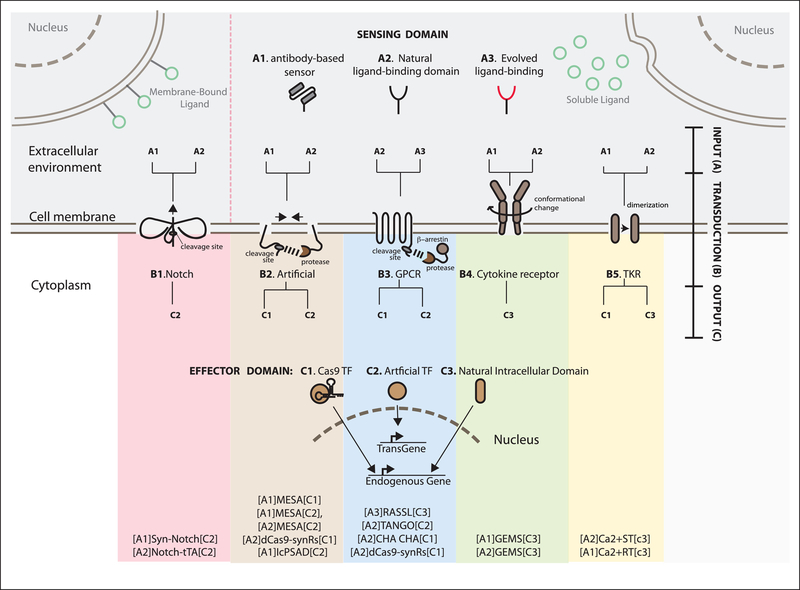
Figure 2 – Mapping existing synthetic receptor systems based on their input, transduction and output characteristics.
A schematic of various synthetic receptors that process information from the extracellular space (top), to inside the cell (bottom).
Sensing domains are classified as antibody-based (A1), based on natural ligand binding domains (A2), or generated via directed evolution (A3). Effector domains are natural (C3) if endogenous domains are used. For transcriptional outputs, we distinguish artificial transcription factors like Gal4 or tTA that activate exogenous expression cassettes (transgene) (C2), or dCas9-based transcription factors that activate endogenous targets (C3). Each sensor and effector domain can be mounted onto a part of the receptor protein that executes the transduction (B). B1 is contact dependent, i.e. is activated only via membrane-bound ligand (green circles presented on a neighboring cell in the upper left corner). B2–5 all recognize ligands that are soluble in the extracellular environment (green circles secreted from other cells in the upper right corner).
At the far bottom, we list the synthetic receptor systems that have been implemented to date using alphanumeric codes according to the notation introduced in the text of [input]transduction[output]. For example, [A1]synNotch[C2] represents the synthetic receptor that uses an antibody-based sensor as input (A1), and activates an artificial transcription factor as output (C2), and so on. The works that describe in the details those receptors are in the references below and in the text.
SynNotch = Synthetic Notch24; MESA = Modular Extracellular Sensor Architecture15,64,65; IcPSAD = Intracellular protein sensor actuator device66; GEMS = generalized extracellular molecule sensor14; RASSL = Receptor activated solely by synthetic ligand17; Tango19; ChaCha20; dCas9 synRs = dCas9 synthetic receptor21; Ca2+ST = calcium sensing-rewiring tool67; Ca2+RT = calcium rewiring tool18,68; GPCR = G-protein coupled receptor; TKR= tyrosine kinase receptor.