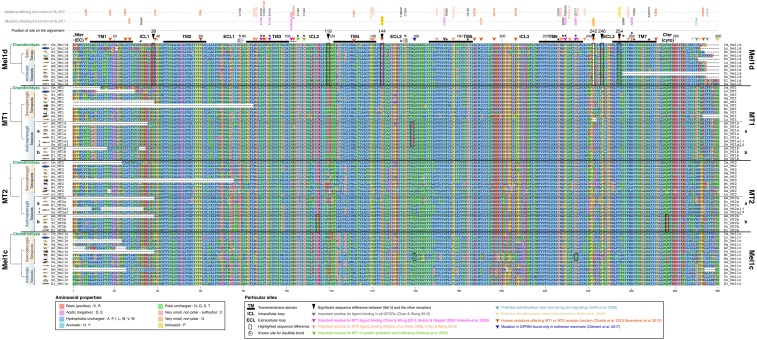
Figure 6.
Alignment used to compare amino acid positions among vertebrate MTR proteins (matching to the alignment used for phylogenetic analysis; Additional Dataset S1). Species abbreviations: Ac = Anolis carolensis (green anole lizard); Am = Astyanax mexicanus (Mexican cavefish); Bt = Bos taurus (cattle); Cm = Callorhinchus milli (elephant shark); Dr = Danio rerio (zebrafish); El = Esox lucius (northern pike); Gg = Gallus gallus (chicken); Hs = Homo sapiens (human); Lc = Latimeria chalumnae (coelacanth); Lo = Lepisosteus oculatus (spotted gar); Oa = Ornithorhynchus anatinus (platypus); On = Oreochromis niloticus (Nile tilapia); Ps = Pelodiscus sinensis (Chinese softshell turtle); Tr = Takifugu rubripes (tiger pufferfish); Xt = Xenopus tropicalis (western clawed frog). Detailed annotation of sequences flagged up in the main text are provided within the figure.