Figure 3.
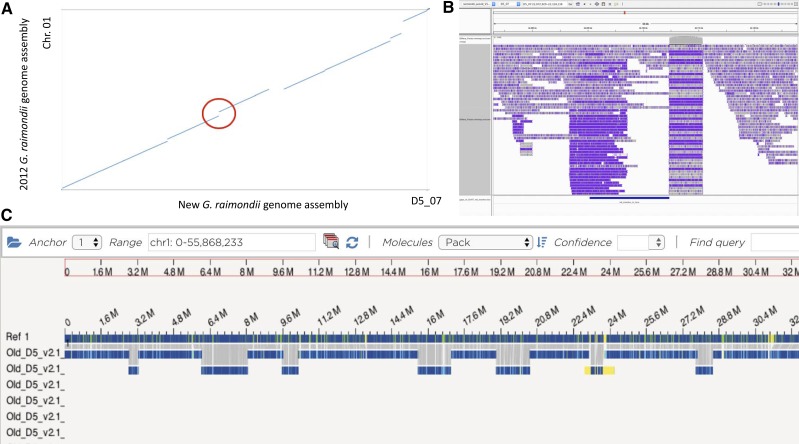
Genomic assembly data of the new G. raimondii sequence suggest that the previously reported mitochondrial insertion was likely due to an assembly error. A) Genome alignments between G. raimondii Chr. 01 (Paterson et al. 2012) and our new genome sequence of D5_07. The red circle indicates the putative position of the mitochondrial genome insertion in the previous G. raimondii sequence relative to the new assembly. B) Alignment of G. raimondii PacBio reads (Track 2) to the new reference genome of G. raimondii (Track 1). The multi-colored bars represent individual PacBio reads (Track 2). The previous reference genome of G. raimondii had a mitochondrial insertion somewhere in this 14kb region indicated by the blue bar of Track 3. There are no PacBio reads that span the gap between the flanking regions of the 6,071 repeat and the repeat itself. C) Bionano data mapped to the previous reference genome sequence of G. raimondii (Paterson et al. 2012) also suggest an insertion of a sequence that is non-contiguous in the flanking regions. The Ref1 track reference to the originally published genome sequence of G. raimondii with a mitochondrial insertion between ∼23Mb and ∼24Mb. Independently constructed Bionano contigs were aligned to the 2012 reference sequence. A Bionano contig matched the reference sequence in the mitochondria insertion region, but the flanking regions of the Bionano contig (yellow) did not match flanking Bionano contigs or the reference sequence.