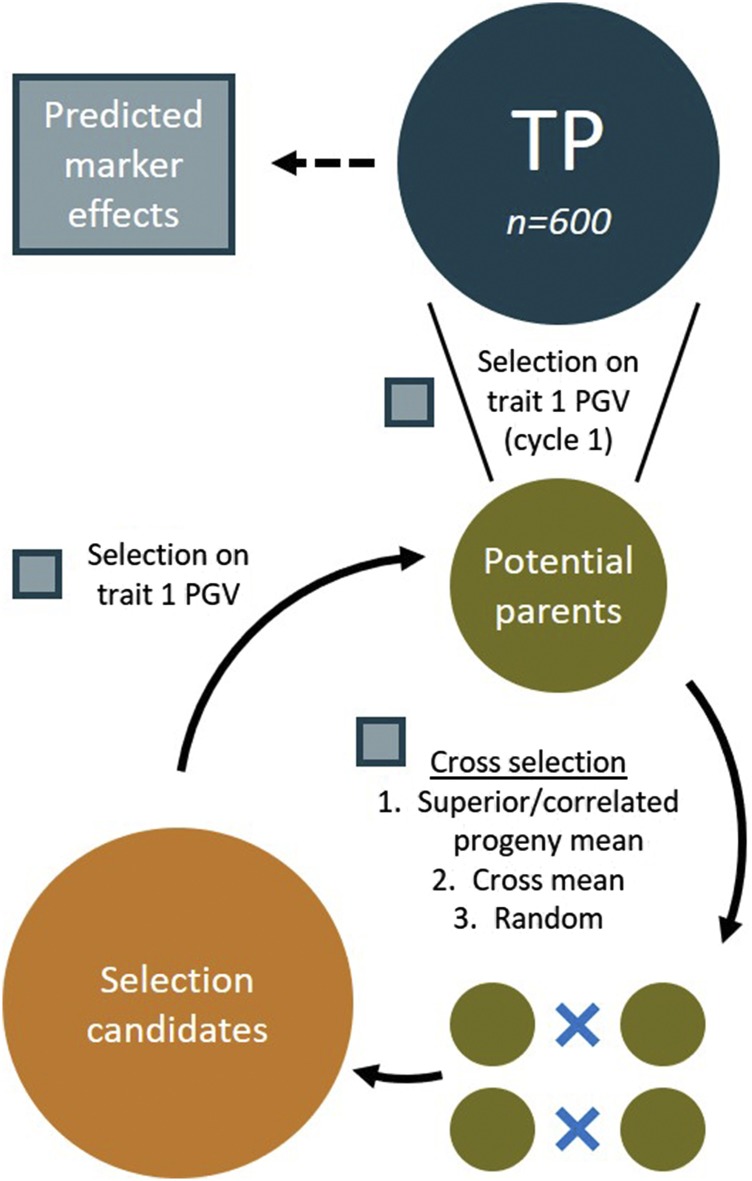
Figure 1.
In the recurrent selection simulation (Simulation 2), a training population (TP) was sampled and used to predict genomewide marker effects. In the first cycle, potential parents were identified using direct selection on predicted genotypic values (PGVs) of the first trait. Crosses were selected by one of three methods and were used to simulate selection candidates. Potential parents of the next cycle were determined using direct selection on the first trait. Ten breeding cycles were simulated. Any processes that relied on the predicted marker effects are noted with a blue/gray box.