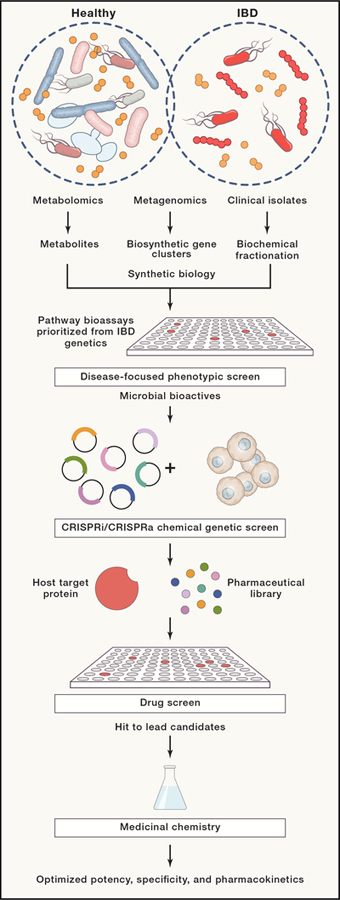
Figure 5. Translating microbiome bioactives into therapeutic agents.
Natural product molecular libraries can be designed and assembled based on microbiome metabolomics datasets. Alternatively, compounds can be directly produced by synthetic biology approaches and/or fractionated from microbial cultures. With microbiome-derived molecular libraries, compounds and molecules can be screened and prioritized based on their bioactivity in phenotypic screening assays that model biological processes and pathways associated with IBD pathology. In turn, chemical-genetic screens can facilitate identification of host target proteins through which microbiome bioactives exert their function. For example, a given microbiome bioactive is predicted to lose its biological effect in host cells that lack their specific target protein due to genetic knock-out or knock-down (CRISPR KO or CRISPRi). Conversely, a given microbiome bioactive is predicted to exhibit increased biological activity in host cells that overexpress their specific target protein (CRISPRa). Subsequently, pharmaceutical libraries with drug-like properties can be screened for activity against validated host targets. Finally, lead candidate small molecules can be further engineered by medicinal chemistry to optimize potency, selectivity, and pharmacokinetic properties.