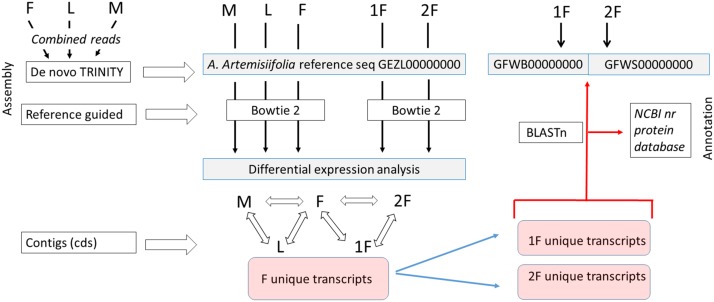
Figure 2. Assembly strategy of A. artemisiifolia floral transcript datasets.
As a first step a de novo Trinity assembly of combined read set of male (M), female (F) and leaf (L) of A. artemisiifolia wild growing plants was performed to get a reference transcriptome that represent both vegetative and generative organ transcripts in different developmental stages. Separately two set of reads representing two classes of F development such as early (1F) and late (2F) stages were generated from sterile in vitro plants. In order to perform differential expression analysis of each five sample, cleaned read sets of M, L, F, 1F, and 2F were assembled by using bowtie2 reference guided algorithm aligning reads to the A. artemisiifolia reference transcriptome. Focusing on the female floral expression pattern in various developmental stages we determined the F unique transcripts collecting the whole female tissue specific contig set. After that 1F and 2F unique transcripts were clustered aligning them to the F contig set. In the second step Trinity de novo assembly were performed using 1F and 2F cleaned reads to get 1F and 2F specific contigs. In order to filter exactly the 1F and 2F specific transcripts, and characteristic transcription pattern to both developmental classes we aligned the 1F and 2F unique transcripts to 1F and 2F de novo Trinity contigs used Blastn algorithm. Thus filtered sequences were annotated using Blastx search and Blast2Go high throughput blast against the NCBI nr protein database. Transcript data sets of de novo Trinity assembly of combined F-L-M as A. artemisiifolia refseq and separately 1F and 2F were deposited in the NCBI TSA database with the accession number shown in the Figure.