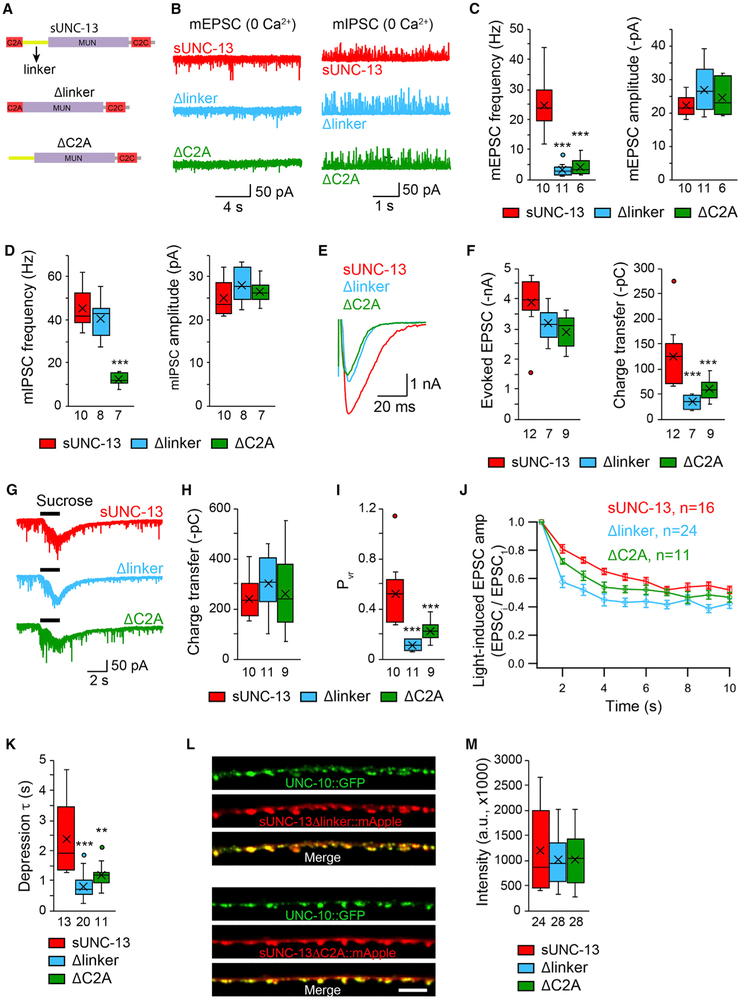
Figure 5. The C2A and Linker Domains Are Essential for sUNC-13 Function.
(A) Cartoon depicting the domain structure of sUNC-13, sUNC-13Δlinker, and sUNC-13ΔC2A.
(B–D) Representative traces and boxplot of the frequency and amplitude of mEPSCs and mIPSCs recorded from sUNC-13 rescue, Δlinker rescue, and ΔC2A rescue animals in 0 mM Ca2+.
(E and F) Representative traces and summary of the amplitude, charge transfer, and decay of the evoked EPSCs recorded from sUNC-13 rescue and Δlinker rescue and ΔC2A rescue in 1 mM Ca2+.
(G–I) Example traces of hypertonic sucrose-evoked currents (G), averaged charge transfer of sucrose currents (H), and Pvr (I) from the indicated genotypes.
(J) 1-Hz light train stimulus-triggered EPSCs from sUNC-13 rescue, Δlinker rescue, and ΔC2A rescue animals.
(K) Synaptic depression, analyzed by the normalization of the amplitude of the EPSCs to the amplitude of the first EPSC.
(L) Representative confocal z stack images for sUNC-13Δlinker and sUNC-13ΔC2A (tagged with mApple) and their colocalization with UNC-10/RIM (tagged with GFP). Scale bar, 5 μm. Bottom: line scans along the dorsal nerve cord.
(M) Quantification of the fluorescence intensity of sUNC-13Δlinker::mApple and sUNC-13ΔC2A::mApple.
Data in (J) are presented as mean ± SEM, and all other data are shown as box-and-whisker plots with median (line) and mean (cross) indicated. **p < 0.01, ***p < 0.001 compared with sUNC-13 rescue; one-way ANOVA test for data in (D), (H), and (M); one-way ANOVA following Kruskal-Wallis test for data in (C), (F), (I), and (K). The number of worms analyzed for each genotype is indicated under each box.