ABSTRACT
Molecular risk stratification of colorectal cancer can improve patient outcome. A panel of lncRNAs (H19, HOTTIP, HULC and MALAT1) derived from serum exosomes of patients with non-metastatic CRC and healthy donors was analyzed. Exosomes from healthy donors carried significantly more H19, HULC and HOTTIP transcripts in comparison to CRC patients. Correlation analysis between lncRNAs and clinical data revealed a statistical significance between low levels of exosomal HOTTIP and poor overall survival. This was confirmed by multivariate analysis that HOTTIP is an independent prognostic marker for overall survival (HR: 4.5, CI: 1.69–11.98, p = 0.0027). Here, HOTTIP poses to be a valid biomarker for patients with a CRC to predict post-surgical survival time.
KEYWORDS: Exosomes, colorectal cancer, diagnosis, liquid biopsy, long non-coding RNA
Introduction
Colorectal cancer (CRC) is one of the most frequent cancers worldwide [1], despite decreasing incidence and mortality. Detection at early stage is the most effective marker for best overall survival. However, most patients present with advanced disease at time of diagnosis and patients with stage 4 disease have a poor 5 year overall survival [2,3]. The current molecular risk classification of CRC is based on histopathology markers such as stage, grade of differentiation and lymph node invasion, which are crucial to assess the tumor initially, but fail to recapitulate the heterogeneity in CRC and its associated wide range of treatment response and overall outcome. There is an urgent clinical need to identify pivotal biomarkers and combine these with the genetic information of a patient’s tumor to improve an accurate prognosis and predict treatment response, risk of progression and overall survival.
Liquid biopsies can be used as a minimal invasive tool to determine malignancies, monitor disease progression and treatment response [4]. Exosomes, commonly found in body fluids, are extracellular vesicles that can contain cell-derived proteins, lipids and nucleic acids including microRNA and long non-coding RNA (lncRNA), and have attracted remarkable interest to detect and monitor diseases [5]. LncRNAs are a subclass of non-protein coding RNAs involved in the gene regulation of cell growth, invasion, and other critical cellular functions [6,7]. They are often expressed in a disease- and tissue-specific manner making these molecules attractive therapeutic targets, and pointing toward specific functions for lncRNAs in development and diseases [8–10]. Recent studies have shown that serum-derived exosomes from cancer patients have a different lncRNA profile in comparison with healthy donors or patients with a non-malignant disease [11–13].
This study aimed to evaluate whether exosomal lncRNAs can be used as minimal-invasive diagnostic and prognostic markers in CRC. Specifically, we investigated the abundance of four lncRNAs that have been associated with the epithelial-to-mesenchymal transition [8,9] and metastasis [8] in serum-derived exosomes from patients with non-metastatic CRC and healthy controls in two independent cohorts.
Materials and methods
Patient characteristics and data collection
Serum collection was approved by the Ethics Committee of the University of Heidelberg and the Ethics Committee of the University of Dresden. Cohort 1 consists of 52 patients with CRC that underwent tumor resection between 2007 and 2009 at the Department of Surgery, University of Heidelberg. Serum samples from 52 healthy donors were included as a control. Cohort 2 consists of 48 patients with primary CRC that underwent tumor resection between 2015 and 2016 at the Department of Surgery, University of Dresden. As controls, serum samples from 24 healthy donors were included. Clinical information was obtained for all patients such as age, gender, TNM classification, grading and treatment with chemotherapy. Overall survival data was available for cohort 1, however outcome data of cohort 2 is currently not available.
Blood collection and isolation of serum exosomes
Blood collection was performed as described recently [11]. Briefly, after blood collection, tubes were immediately centrifuged at 2500g for 10min, and separated serum was stored at −80°C. For exosome isolation, serum samples were thawed on ice. Subsequently, exosomes were isolated using an exosome isolation kit (ThermoFisher Scientific®, Cat-Nr.: 4478360) following the manufacturer’s instructions. Exosome validation included nanoparticle tracking analysis (ZetaView®, Particle Metrix GmbH) and immunoblotting of exosomal markers.
Nanoparticle tracking analysis including fluorescent mode by Zetaview®
Nanoparticle tracking analysis (NTA) was conducted using ZetaView® to determine the concentration and size of the extracted exosomes according to the manufacturer’s instructions. In addition, the fluorescent mode of ZetaView® was employed to measure exosomal markers on the surface of the extracted extracellular vesicles. For this analysis, 6.6*10 [9] exosomes were resuspended in 100µl 1xPBS and incubated with 0.3µm polystyrene latex beads (Sigma-Aldrich, Cat.Nr.: LB30–1ML) on a horizontal shaker with 1000rpm for 15min. Subsequently, this suspension was diluted to 1ml with PBS and incubated on a horizontal shaker with 600rpm for 60min. For blocking, 100mM glycine and 1% BSA in PBS was added and incubated for 30min. The suspension was centrifuged at 14.000g for 3min, and the bead-bound exosomes were recovered as a pellet. The pellet was diluted in 40µl PBS and divided into two equal volumes. One aliquot was incubated with primary antibodies (CD81 polyclonal rabbit, dilution 1:20, Novus Biologicals, Cat.Nr.: NBP2–20564; CD63 polyclonal rabbit, dilution 1:20, Santa Cruz, Cat.Nr. sc-15363; CD9 monoclonal rabbit, dilution 1:20, Abcam, Cat.Nr.: ab92726; TSG101, monoclonal mouse, dilution: 1:100, Abcam, Cat.Nr.: ab83), the second aliquot was used as negative control. After 1hr of incubation, the reaction was stopped by adding blocking solution. Subsequently, the suspension was centrifuged at 14.000g for 3min, and the bead-bound exosomes were recovered as a pellet. The Alexa-488 secondary antibody (Life Technologies, NY 14,072, USA, anti-rabbit: A11034 or anti-mouse: A11029) was added to all aliquots and incubated for 60min. After washing with 1x PBS/2% BSA, the pellet was diluted in 1ml 1x PBS. The fluorescent bead-bound exosomes (positive staining and negative control) were analyzed using ZetaView® and ZetaView® 8.03.04.01 Software.
Transmission electron microscopy (TEM)
Fixed exosome specimens at an optimal concentration were placed onto 400-mesh carbon/formvar-coated grids and allowed to absorb to the formvar for a minimum of 1min. After rinsing in 1x PBS followed by distilled water, the grids were allowed to dry and stained for contrast using uranyl acetate. The samples were viewed with a Tecnai Bio Twin transmission electron microscope (FEI), and images were taken with an AMT CCD Camera (Advanced Microscopy Techniques).
Immunoblotting
Immunoblot analysis was performed as described recently with some minor modifications [11]. Briefly, serum exosomes were harvested in RIPA buffer including Halt™ Protease- and Phosphatase-Inhibitor-Cocktail (Thermo Scientific). 20µg of protein were loaded and analyzed using acrylamide gels. Subsequently, proteins were transferred onto PVDF membranes (ImmobilonP) and blocked with 5% non-fat dry milk or 5% BSA in PBS/0,05% Tween for 1hr. Subsequently, the blot was incubated with primary antibodies against CD9 (dilution 1:500), TSG101 (dilution 1:200) and CD63 (dilution 1:200) at 4°C overnight. After incubation with horseradish peroxidase coupled secondary antibodies, blots were developed with chemiluminescent reagent and visualized using an Imagers G:Box Chemi XT4 (Syngene).
RNA isolation and real-time qPCR
Total RNA from exosomes was extracted by employing the Total Exosome RNA & Protein Isolation Kit (Invitrogen) following the manual´s instructions. RNA quality and concentration were evaluated using a NanoVueTM Plus spectral photometer (GE Healthcare BioSciences). Total RNA was reversely transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Fifty nanograms of the resulting cDNA were further used for quantification by qPCR (SYBR Green PCR Kit, Qiagen, Hilden, Germany) in a Step One Plus System (Applied Biosystems). Following primers were used: H19 Forward: 5′-GCACCTTGGACATCTGGAGT-3′, Reverse: 5′-TTCTTTCCAGCCCTAGCTCA-3′; HOTTIP Forward: 5′-CCTAAAGCCACGCTTCTTTG-3′, Reverse: 5′-TGCAGGCTGGAGATCCTACT-3′[14]; HULC: Forward: 5′-TCATGATGGAATTGGAGCCTT-3′, Reverse: 5′-CTCTTCCTGGCTTGCAGATTG-3′[15]; MALAT1 Forward: 5′-CGTCATTTAAAGCCTAGTTAACGCA-3′, Reverse: 5′-GTTTCATCCTACCACTCCCAA-3′. Samples were normalized to 28s RNA (Forward: 5′-AACGAGATTCCCACTGTCCC-3′, Reverse: 5′-CTTCACCGTGCCAGACTAGAG-3′).
Statistical analysis
The SPSS software package version 21.0 (Chicago, IL, USA) Graph Pad and MedCalc were used for statistical analysis. Mann-Whitney-U test was performed to compare the expression of lncRNAs in serum exosomes. Chi-squared (χ[2]) tests were applied to examine the correlation between the expression of exosomal lncRNAs and clinicopathological parameters. Receiver operating characteristic (ROC) curves were used to determine the sensitivity and specificity, and to compare the area under the curves (AUC) of exosomal lncRNAs. Univariate survival analysis was performed using the Kaplan-Meier method and differences in survival were calculated with the log-rank test whilst the Cox proportional hazards regression model was conducted for multivariate testing. Results were considered significant at a p-value less than 0.05.
Results
Patient characteristics
To conduct the study, we used cohort 1 consisting of 52 patients with CRC and 52 healthy donors from primary CRC patients treated in Heidelberg, Germany. The second cohort (cohort 2) was used to validate our findings and consisted of 48 patients with CRC and 24 healthy donors. The clinicopathological and patient characteristics have been summarized in Table 1.
Table 1.
Univariate analysis (log-rank test) of prognostic parameters in primary colorectal cancer of cohort 1 for overall survival. Data was missing for some patients regarding parameters *Grading (n = 39 total), **Resection status (n = 47 total) and ***Neoadjuvant therapy (n = 28).
| Prognostic parameters | Number of patients (n=) | Mean overall survival (months) | P-Value |
|---|---|---|---|
| Age | 0.1573 | ||
| < median | 26 | 75.35 | |
| ≥ median | 26 | 67.05 | |
| Gender | 0.9214 | ||
| Female | 19 | 68.93 | |
| Male | 33 | 72.36 | |
| Localization of the tumor | 0.1422 | ||
| Rectum | 39 | 76.45 | |
| Colon | 13 | 57.59 | |
| UICC stage | |||
| I/II | 28 | 78.69 | 0.0735 |
| III/IV | 24 | 61.67 | |
| Lymph node metastasis | |||
| negative | 30 | 79.79 | 0.0264 |
| positive | 22 | 58.55 | |
| Grading* | 0.6954 | ||
| 1/2 | 33 | 69.99 | |
| 3/4 | 6 | 62.48 | |
| Resection status** | 0.7578 | ||
| R0 | 43 | 71.71 | |
| R1 | 4 | 68.97 | |
| Neoadjuvant therapy*** | 0.5671 | ||
| Received | 25 | 73.67 | |
| Not received | 3 | 68.78 | |
| Exosomal lncRNA HOTTIP | 0.0009 | ||
| Low/intermediate expression | 39 | 47.00 | |
| High expression | 13 | 80.42 |
Identification of serum exosomes
Firstly, we sought out to determine the existence of exosomes in the serum from patients with CRC and healthy donors. After extraction, extracellular vesicles (EV) were measured using ZetaView® with a median size of 109.3 nm/EVs ranging from 73.2 – 136nm (Figure 1A). TEM confirmed the presence of double-membrane vesicles with a size range of 50–150nm (Figure 1B). Isolated EVs were positive for common exosome markers [16] CD9, CD81, CD63 and TSG 101 using immunoblot analyses (Figure 1C), and calreticulin was used to control for potential cytoplasmic contamination, which was hardly expressed in our samples. Fluorescent staining of TSG 101, CD9, CD81 and C63 revealed that bead-bound EVs were strongly positive for these exosomal markers (Figure 1D). Taken together, we detected classical exosomes [16] with a diameter of 50 – 150 nm enriched of exosome-related proteins such as CD9, CD63, CD81 or TSG 101 in the serum of healthy donors and patients with underlying diseases [17,18].
Figure 1.
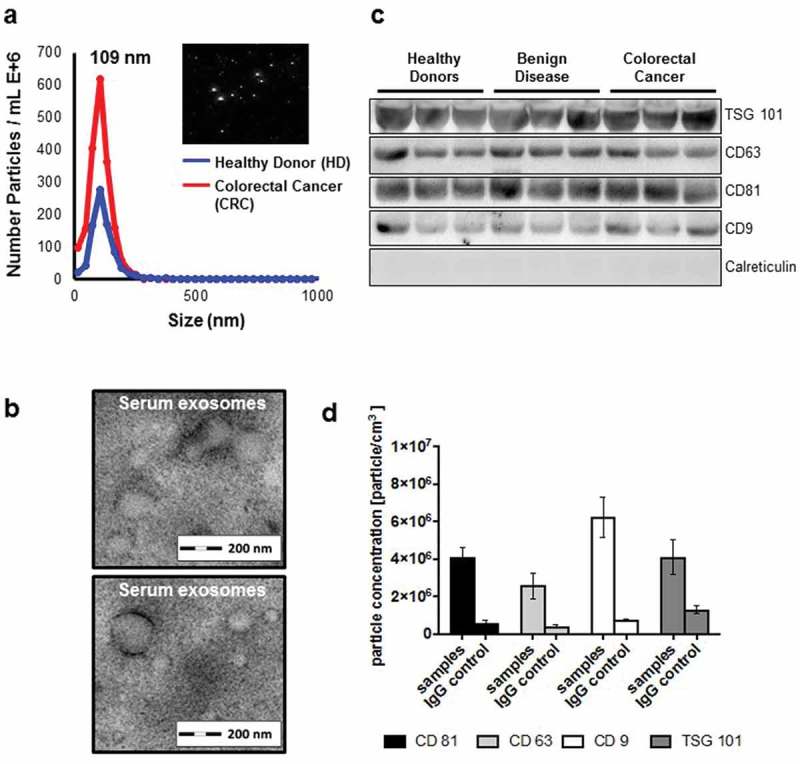
(a) Identification of serum exosomes by nanoparticle tracking analysis (NTA) using ZetaView®, (b) by transmission electron microscopy, (c) by Western Blot for exosome-associated proteins and (d) by fluorescent NTA for exosome-associated protein markers.
Expression profile of exosomal long non-coding RNAs H19, HOTTIP, HULC and MALAT in serum-derived exosomes
Long non-coding RNAs (lncRNA) have been implicated to contribute to the onset and progression of various diseases [8], as these are known modulators of gene expression. We focused specifically on four lncRNA (H19, HOTTIP, MALAT1 and HULC) that have been associated with CRC [19–25]. HOTTIP, MALAT1 and H19 are aberrantly expressed in CRC and have been associated with a worse prognosis and poor overall survival. These lncRNAs are known regulators of the EMT transition, as they are involved in the repression of tumor suppressive microRNAs, epigenetic regulation of HOXA and EZH2 genes and the remodeling of the chromatin complexes [26].
At first we wanted to investigate whether these lncRNA can be detected in the serum-derived exosomes and found that lncRNA H19, HOTTIP and HULC expression was significantly decreased in serum-derived exosomes from CRC patients compared to healthy donors (Figure 2A–C). In contrast, no significant difference was observed of exosome-derived MALAT1 between healthy donors and patients with CRC (Figure 2D). To validate our findings, we used a second cohort of primary CRC and healthy donors available to us. We found a similar trend of the lncRNAs from cohort 1, suggesting that exosomal HOTTIP, HULC and H19 are downregulated in primary CRC patients. Next, we asked whether these lncRNAs are predictive of disease and performed ROC curve analysis for all lncRNAs in both cohorts (Figure 3A, B). Exosomal HOTTIP in healthy donors vs. cancer patients resulted in an area under the curve (AUC) of 0.75 in cohorts 1 and 2 (Figure 3A, B, Supplementary Table 1). The AUC of exosomal HULC for healthy donors vs. cancer patients reached 0.77 (cohort 1) and 0.71 (cohort 2) (Figure 3A, Supplementary Table 1). For H19, ROC curve analysis showed an AUC value of 0.68 (cohort 1) and an AUC of 0.72 (cohort 2) in healthy donors vs. cancer patients (Figure 3A, Supplementary Table 1), whilst exosomal MALAT1 showed the lowest AUC of 0.61 and 0.62 in cohort 1 and cohort 2.
Figure 2.
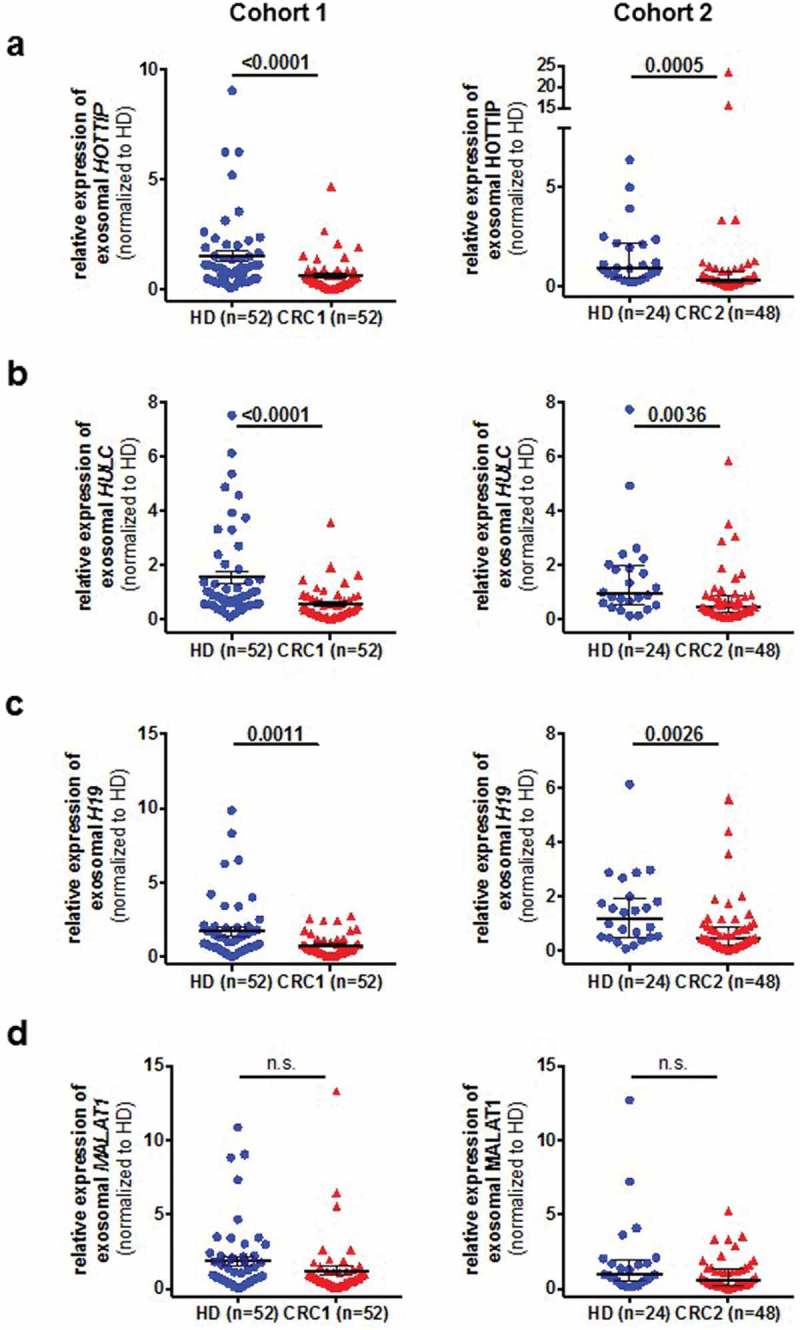
Expression analysis of exosomal long non-coding RNAs H19 (A), HOTTIP (B), HULC (C) and MALAT1 (D) in two independent cohorts (cohort 1, left panels, cohort 2, right panels) in specimens from healthy control (HC) and patients with colorectal cancer (CRC). Expression level is indicated by the normalized Ct-values.
Figure 3.
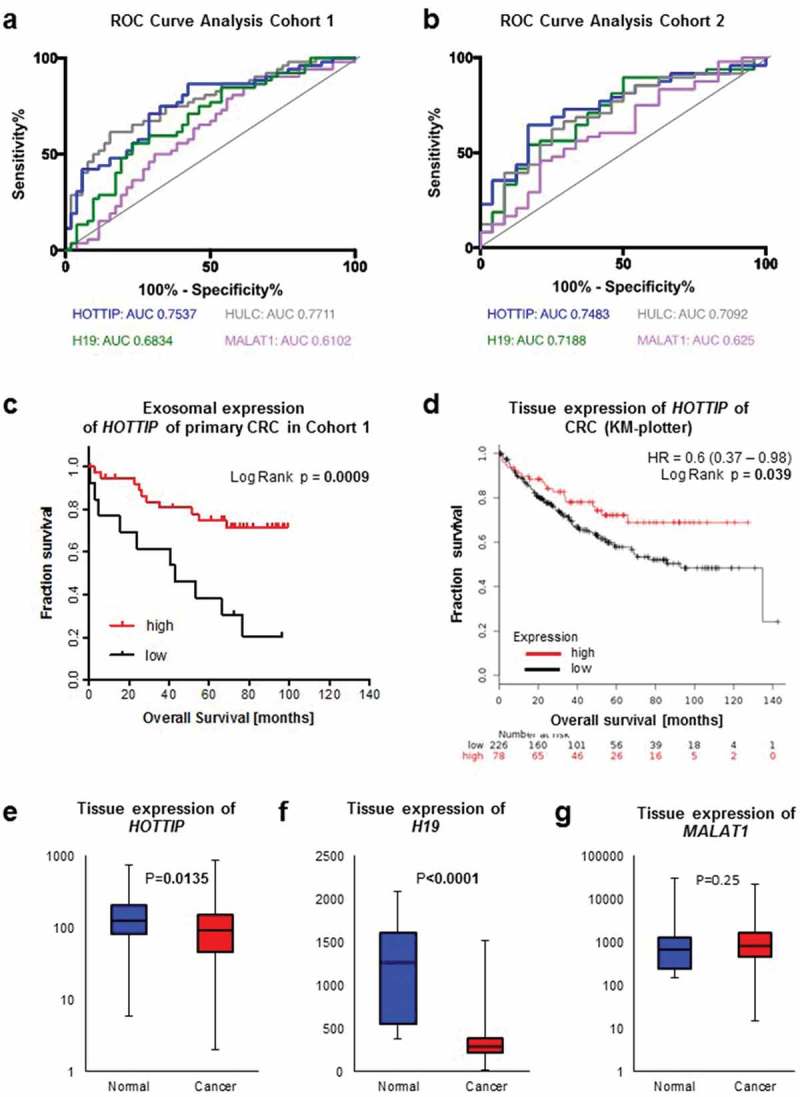
ROC curve analysis of exosomal long non-coding RNAs H19 (green), HOTTIP (blue), HULC (grey) and MALAT1 (violet) from patients with colorectal cancer and healthy control (a) of cohort 1 and (b) cohort 2. (c) Kaplan-Meier analysis of overall survival in patients with colorectal cancer expressing high (red line) or low (black line) serum-derived exosomal HOTTIP. (d) Kaplan-Meier analysis of overall survival in patients with colorectal cancer and high (red line) or low (black line) expression of HOTTIP in primary colorectal cancer. Data was generated using KM-plotter. (e-g) Expression analysis of (e) HOTTIP, (f) H19 and (g) MALAT1 in colorectal cancer tissue or normal tissue. Data was generated using KM-plotter.
Low expression of HOTTIP in exosomes and tissue of primary CRC is a prognostic marker for decreased overall survival
Next, we asked whether the levels of HOTTIP expression might be indicative of patient outcome in our cohort 1 as exosomal HOTTIP, HULC and H19 expression resulted in a good differential between health and disease. For survival analysis, we categorized the patients accordingly into low/intermediate (<75th percentile) and high (≥75th percentile) expression of exosomal lncRNA. Chi-square analysis revealed no significant correlation between any histopathological parameter and the exosomal expression of HOTTIP, HULC or H19 (data not shown). Univariate analysis by log-rank tests showed a highly significant association between low/intermediate expression of exosome-derived HOTTIP and poor overall survival (Low/intermediate: 47.0 months, High: 80.4 months, p = 0.0009) (Figure 3C, Table 1), whilst no statistical significance in overall survival was observed depending on H19 and HULC expression (Supplementary Figure 1A, 1B). In addition, the number of positive lymph nodes (p = 0.026) was a significant prognostic marker (Table 1, Supplementary Figure 1C). Multivariate analysis confirmed low expression of HOTTIP as an independent prognostic marker for overall survival (HR: 4.5, CI: 1.69–11.98, p = 0.0027), whereas the lymph node status failed to be significant (Table 2). Next, we assessed the expression of HOTTIP, H19 and MALAT1 in an independent cohort of patients with CRC [27] to confirm our findings. Of note, overall survival data was not available for HULC. In concordance with the exosomal expression, low/intermediate expression of HOTTIP in primary CRC tissue was an indicator for poor survival (Figure 3D) (HR: 0.6, CI: 0.37–0.98, p = 0.039) in a cohort of 308 patients with CRC. We also found a significantly lower expression of HOTTIP and H19 in these tumor tissues (n = 2110) compared to normal tissue (n = 40) (p < 0.05, respectively, Figure 3E, 3F) whilst no significant difference in the expression of MALAT1 was observed between normal and cancerous tissue (p = 0.25) (Figure 3G). Taken together, we were able to show that lncRNAs of exosomes can be used as a diagnostic marker.
Table 2.
Multivariate analysis (Cox proportional hazards regression model) of prognostic parameters for overall survival in colorectal cancer.
| Characteristics | Hazard Ratio | 95% CI of Relative Risk | P-Value |
|---|---|---|---|
| Exosomal lncRNA HOTTIP(Low/intermediate vs high) | 4.494 | 1.685–11.984 | 0.0027 |
| Age (< median vs. ≥ median) | 1.581 | 0.591–4.229 | 0.3618 |
| Gender (female vs. male) | 0.638 | 0.215–1.890 | 0.4169 |
| pT-Stage (pT 1/2 vs. pT 3/4) | 2.496 | 0.565–11.030 | 0.2275 |
| Lymph node metastasis (positive vs negative) | 1.776 | 0.642–4.912 | 0.2686 |
Discussion
Molecular risk stratification in colorectal cancer is based on histopathology markers such as stage, poor differentiation and lymph node invasion, which currently fails to predict which patient with local or advanced CRC will benefit from post-surgical treatments with minimal adverse events or complications. This urgent clinical need could be met by using circulating components of the blood as an attractive tool to discover highly specific and non-invasive biomarkers. Recent studies have shown the relevance of exosomal-associated protein signatures and microRNAs in various diseases; hence the use of exosomal lncRNAs as clinical biomarkers is still undergoing. Therefore, we assessed a panel of four prominent lncRNAs that have been described to play a pivotal role in CRC progression and dissemination [22,25,28–30]. In concordance with REMARK [31], we evaluated the presence of these four selected candidate lncRNAs in two independent cohorts. Both cohorts revealed that H19, HULC and HOTTIP are decreased in exosomes derived from serum of patients suffering from CRC. For exosomal H19, these results have been observed in an independent dataset (www.exorbase.org), as patients with CRC have lower H19 expression in exosomes compared to healthy volunteers or patients with coronary heart disease. Moreover, we show a concordant decreased expression of H19 and HOTTIP in tumor tissue and exosomes from cancer patients. In addition, we have not observed any differential expression of MALAT1 in tissue and exosomes from patients with CRC and control. These data underline the hypothesis that tumor-derived exosomal cargo in the blood circulation can reflect the expression profile of their cells of origin. However, these results are contradicting the pro-tumorigenic effects of H19 and HOTTIP in CRC that have been reported so far [25,28]. The patient cohorts were mostly primary CRC of the colon with no information about presence of metastasis or resection status [25,28], whereby our cohorts included mostly patients with non-metastatic CRC of the rectum that underwent R0 resection. In addition, the biogenesis of exosomes is a selective enrichment process by which the parental cell could regulate the intracellular expression profile of distinct genes [32]. This implies that decreased secretion of exosomal lncRNAs may result in an intracellular accumulation of these pro-tumorigenic biomolecules. One limitation of our study is that the exosomal lncRNAs could originate either from the tumor cells or from other cells, e.g., platelets or leukocytes [32]. The current extractions are unselective, with no indication from which distinct cell populations the exosomes derived and can therefore significantly influence the results. In order to address this hypothesis, it would be necessary to selectively enrich tumor exosomes from the blood using specific surface markers prior to the expression analysis of lncRNAs such as HOTTIP. Another limiting factor of this study is the patient annotation for cohort 1 and outcome data for cohort 2. We were not able to correlate our findings to the genetic composition of the CRC patients from cohort 1 as the KRAS/BRAF and microsatellite stability status was not established. Cohort 2, which is a prospective study, had no overall survival data available at the time of submission.
Moreover, due to the small sample size of serum exosomes, we have to consider the possibility that the retrospective cohort does not approximate the true model for the entire population, suggesting a relative risk of overfitting, which may have caused a bias of the results of our multivariate analysis. This might explain the non-significant association of overall survival with traditional risk factors including pT-stage. To address such bias, additional prospective translational multi-center studies are needed to confirm HOTTIP as a non-invasive prognostic marker in colorectal cancer in the near future.
Overall, our study provides new insights into diagnostic of exosomes and its enclosed lncRNA cargo for patients with CRC. We show that the lncRNA HOTTIP has not only the potential to identify patients with CRC but could serve as a valuable surrogate biomarker for pre-surgical risk stratification as HOTTIP expression significantly predicted the survival time after surgery. This warrants further studies to (a) determine the diagnostic implications by combining established CRC markers and the expression of exosomal HOTTIP in patients suspected to suffer from CRC and (b) investigate the change of expression of lncRNA during disease progression and in response to therapy.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft [KA 3511/3–1]; Nemzeti Kutatási Fejlesztési és Innovációs Hivatal (NKFIH) [NVKP_16–1–2016–0037]; Nemzeti Kutatási Fejlesztési és Innovációs Hivatal (NKFIH) [KH-129581]; Roland Ernst Foundation [5/15].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary data
Suppementary data for this article can be accessed here.
References
- [1].Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16:201–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nordlinger B, Sorbye H, Glimelius B, et al. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14:1208–1215. [DOI] [PubMed] [Google Scholar]
- [4].Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. [DOI] [PubMed] [Google Scholar]
- [5].Xu R, Rai A, Chen M, et al. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15:617–638. [DOI] [PubMed] [Google Scholar]
- [6].Jacob R, Zander S, Gutschner T. The dark side of the epitranscriptome: chemical modifications in long non-coding RNAs. Int J Mol Sci. 2017;18:2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Richtig G, Ehall B, Richtig E, et al. Function and clinical implications of long non-coding RNAs in melanoma. Int J Mol Sci. 2017;18:715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Conigliaro A, Costa V, Lo Dico A, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Barbagallo C, Brex D, Caponnetto A, et al. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Ther Nucleic Acids. 2018;12:229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tang L, Zhang W, Su B, et al. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed Res Int. 2013;2013:251098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jeppesen DK, Fenix AM, Franklin JL, et al. Reassessment of exosome composition. Cell. 2019;177:428–45 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. [DOI] [PubMed] [Google Scholar]
- [18].Lotvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the international society for extracellular vesicles. J Extracell Vesicles. 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Han D, Gao X, Wang M, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159–22173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Klec C, Gutschner T, Panzitt K, et al. Involvement of long non-coding RNA HULC (highly up-regulated in liver cancer) in pathogenesis and implications for therapeutic intervention. Expert Opin Ther Targets. 2019;23:177–186. [DOI] [PubMed] [Google Scholar]
- [22].Yang XJ, Huang CQ, Peng CW, et al. Long noncoding RNA HULC promotes colorectal carcinoma progression through epigenetically repressing NKD2 expression. Gene. 2016;592:172–178. [DOI] [PubMed] [Google Scholar]
- [23].Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–3181. [PMC free article] [PubMed] [Google Scholar]
- [24].Ji Q, Cai G, Liu X, et al. MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis. Cell Death Dis. 2019;10:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ren YK, Xiao Y, Wan XB, et al. Association of long non-coding RNA HOTTIP with progression and prognosis in colorectal cancer. Int J Clin Exp Pathol. 2015;8:11458–11463. [PMC free article] [PubMed] [Google Scholar]
- [26].Gugnoni M, Ciarrocchi A. Long noncoding RNA and epithelial mesenchymal transition in cancer. Int J Mol Sci. 2019;20:1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sztupinszki Z, Gyorffy B. Colon cancer subtypes: concordance, effect on survival and selection of the most representative preclinical models. Sci Rep. 2016;6:37169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lian Y, Ding J, Zhang Z, et al. The long noncoding RNA HOXA transcript at the distal tip promotes colorectal cancer growth partially via silencing of p21 expression. Tumour Biol. 2016;37:7431–7440. [DOI] [PubMed] [Google Scholar]
- [29].Chen SW, Zhu J, Ma J, et al. Overexpression of long non-coding RNA H19 is associated with unfavorable prognosis in patients with colorectal cancer and increased proliferation and migration in colon cancer cells. Oncol Lett. 2017;14:2446–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wu C, Zhu X, Tao K, et al. MALAT1 promotes the colorectal cancer malignancy by increasing DCP1A expression and miR203 downregulation. Mol Carcinog. 2018;57:1421–1431. [DOI] [PubMed] [Google Scholar]
- [31].Sauerbrei W, Taube SE, McShane LM, et al. Reporting recommendations for tumor marker prognostic studies (REMARK): an abridged explanation and elaboration. J Natl Cancer Inst. 2018;110:803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl). 2013;91:431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


