Figure 2.
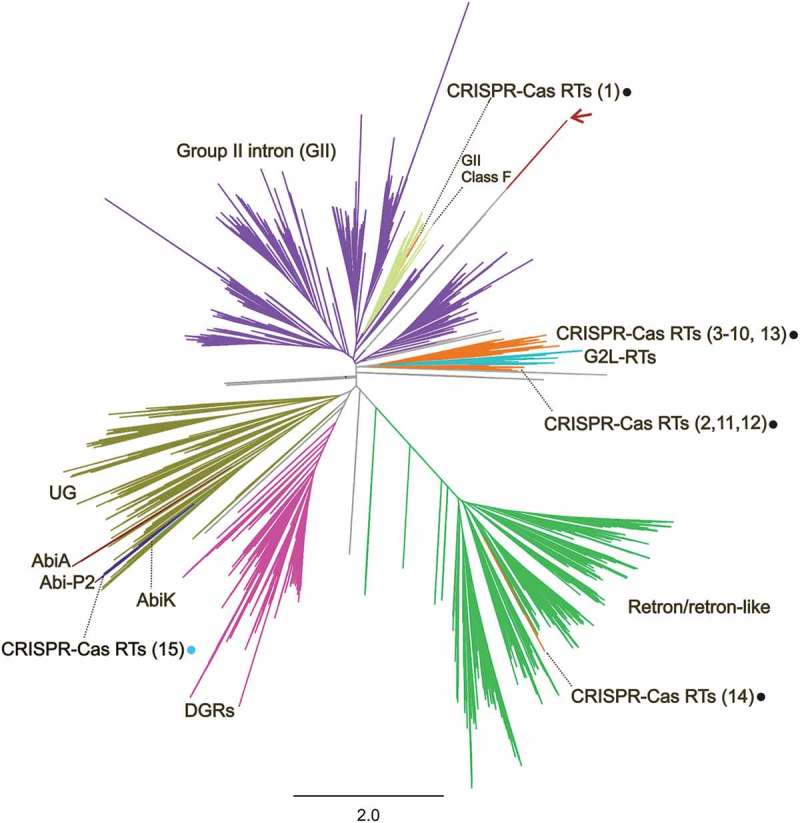
Phylogeny of prokaryotic RTs. The unrooted tree was constructed from an alignment of 9,141 unique predicted RT protein sequences obtained with the FastTree program. The corresponding RT protein sequences, accession number and species names are provided in Supplementary Table 1 and the tree newick file is provided as Supplementary File 1. The branches corresponding to group-II introns (GII), GII class F, Retron/retron-like, DGRs, CRISPR-Cas, G2L, Abi and UG RTs are indicated and highlighted with distinct colours. The numbers of the CRISPR-Cas encoded RT clades are indicated in brackets and the dots indicate the type of system with which they are associated: type III (black) or type I-C (blue). The red arrow indicates the branches corresponding to the putative RTs linked to type I-E CRISPR-Cas systems described by Silas et al. [16]. Relevant subtrees are provided in Figure 4 and Supplementaries Figures 2–4.
