Figure 2.
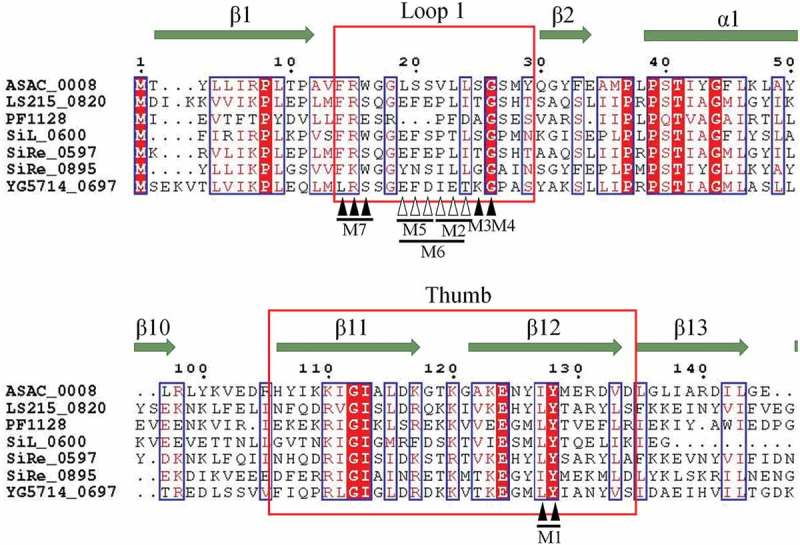
Sequence alignment of Cmr3α homologs using ESPript 3.x (http://espript.ibcp.fr) [50]. Secondary structure is shown above the alignment, based on the structure of Pyrococcus furiosus Cmr3 (PDB: 3X1L, Chain B). Sequences for the alignment are from Acidilobus saccharovorans 345–15 (ASAC_0008), Sulfolobus islandicus L.S.2.15 (LS215_0820), Pyrococcus furiosus DSM 3638 (PF1128), Sulfolobus islandicus LAL14/1 (SiL_0600), Sulfolobus islandicus REY15A (SiRe_0597, SiRe_0895) and Sulfolobus islandicus Y.G.57.14 (YG5714_0697). Shown are sequences containing loop 1 and thumb regions, which are highlighted in red box. The residues marked with black arrows are chosen for substitution mutation (M1: I123A-Y124A; M3: G24F; M4: G25F; M7: F13A-K14A-W15A), while those marked with white arrows are chosen for deletion mutation (M2: deletion of I21-L22-L23; M5: deletion of Y18-N19-S20; M6: deletion from Y18 to L23).
